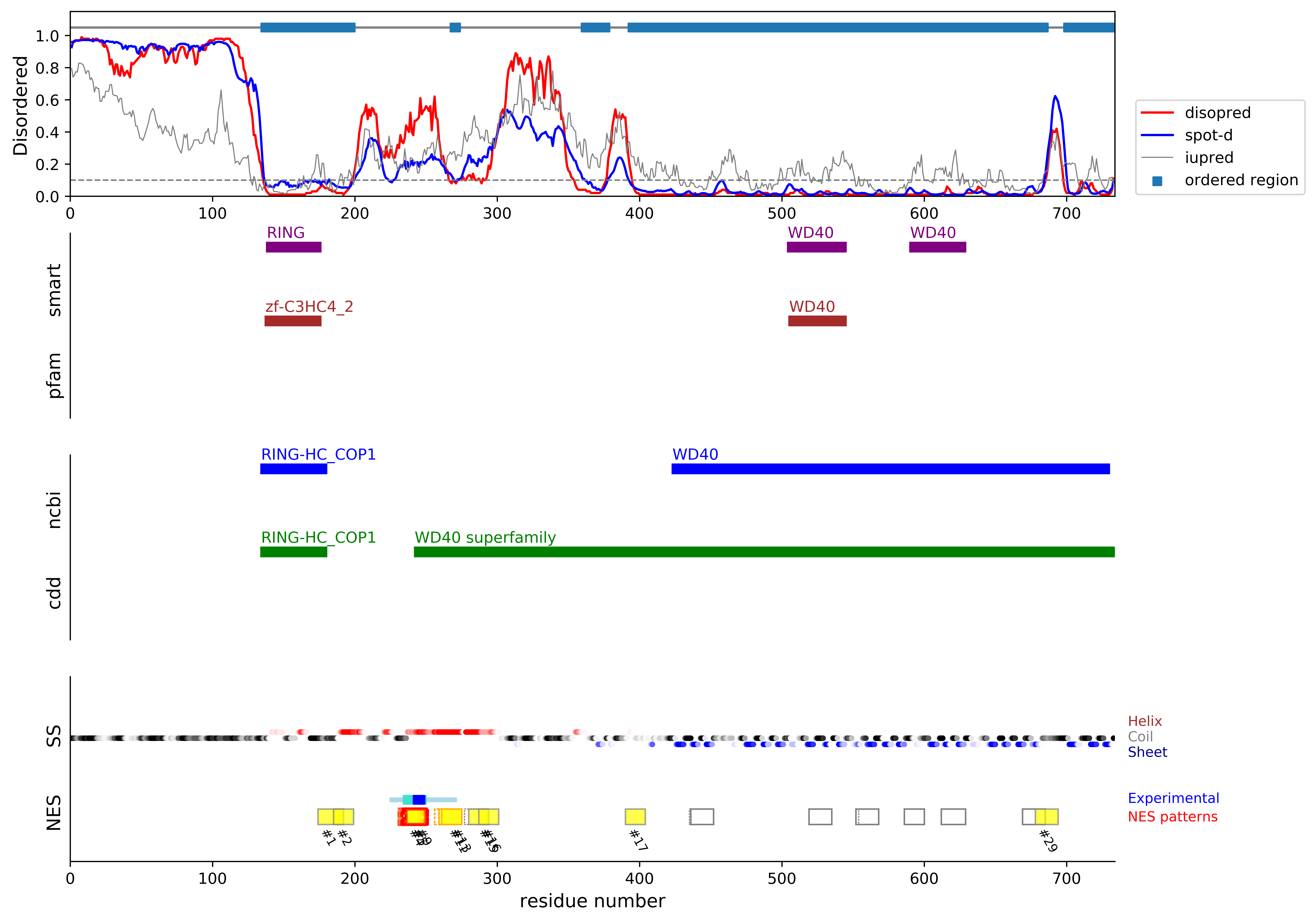

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9R1A8 | 174 | KCNYVVDNIDHLYPNFLV | CCCCCCCCCCCCCCCHHH | c4-4 | uniq | 0.039 | 0.077 | 0.107 | boundary | 0.0 | |
| 2 | fp_D | Q9R1A8 | 185 | LYPNFLVNELILKQ | CCCCHHHHHHHHHH | c2-4 | uniq | 0.036 | 0.06 | 0.099 | boundary | 0.0 | |
| 3 | cand_D | Q9R1A8 | 231 | DQDNLDLANVNLMLEL ......+++++++*+* | CCCCCCHHHHHHHHHH | c1d-AT-4 | multi | 0.424 | 0.198 | 0.198 | DISO | boundary|WD40 superfamily; | 0.0 |
| 4 | cand_D | Q9R1A8 | 231 | DQDNLDLANVNLML ......+++++++* | CCCCCCHHHHHHHH | c2-4 | multi-selected | 0.406 | 0.198 | 0.203 | DISO | boundary|WD40 superfamily; | 0.0 |
| 5 | cand_D | Q9R1A8 | 233 | DNLDLANVNLMLELLV ....+++++++*+*+. | CCCCHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.447 | 0.202 | 0.203 | DISO | boundary|WD40 superfamily; | 0.0 |
| 6 | cand_D | Q9R1A8 | 233 | DNLDLANVNLMLELLV ....+++++++*+*+. | CCCCHHHHHHHHHHHH | c1d-4 | multi | 0.447 | 0.202 | 0.203 | DISO | boundary|WD40 superfamily; | 0.0 |
| 7 | cand_D | Q9R1A8 | 234 | NLDLANVNLMLELLVQK ...+++++++*+*+... | CCCHHHHHHHHHHHHHH | c1c-AT-5 | multi | 0.465 | 0.203 | 0.213 | DISO | boundary|WD40 superfamily; | 0.0 |
| 8 | cand_D | Q9R1A8 | 236 | DLANVNLMLELLVQK .+++++++*+*+... | CHHHHHHHHHHHHHH | c1b-5 | multi | 0.481 | 0.203 | 0.21 | DISO | boundary|WD40 superfamily; | 0.0 |
| 9 | cand_D | Q9R1A8 | 236 | DLANVNLMLELLVQ .+++++++*+*+.. | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.476 | 0.202 | 0.206 | DISO | boundary|WD40 superfamily; | 0.0 |
| 10 | fp_D | Q9R1A8 | 256 | AESHAAQLQILMEFLK ............... | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.237 | 0.17 | 0.263 | DISO | boundary|WD40 superfamily; | 0.0 |
| 11 | fp_D | Q9R1A8 | 259 | HAAQLQILMEFLKVAR ............ | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.158 | 0.146 | 0.253 | DISO | boundary|WD40 superfamily; | 0.0 |
| 12 | fp_D | Q9R1A8 | 259 | HAAQLQILMEFLKVAR ............ | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.158 | 0.146 | 0.253 | DISO | boundary|WD40 superfamily; | 0.0 |
| 13 | fp_D | Q9R1A8 | 261 | AQLQILMEFLKVAR .......... | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.134 | 0.135 | 0.246 | DISO | boundary|WD40 superfamily; | 0.0 |
| 14 | fp_D | Q9R1A8 | 277 | KREQLEQIQKELSVLE | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.109 | 0.221 | 0.396 | DISO | MID|WD40 superfamily; | 0.0 |
| 15 | fp_D | Q9R1A8 | 280 | QLEQIQKELSVLEE | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.111 | 0.228 | 0.404 | DISO | MID|WD40 superfamily; | 0.0 |
| 16 | fp_D | Q9R1A8 | 287 | ELSVLEEDIKRVEE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.191 | 0.282 | 0.36 | DISO | MID|WD40 superfamily; | 0.0 |
| 17 | fp_D | Q9R1A8 | 390 | QLDEFQECLSKFTR | CCHHHHHCCCCCCC | c3-4 | uniq | 0.121 | 0.07 | 0.24 | boundary | MID|WD40 superfamily; | 0.0 |
| 18 | fp_O | Q9R1A8 | 435 | CDYFAIAGVTKKIKVYE | CCEEEEECCCCCEEEEE | c1c-AT-4 | multi | 0.011 | 0.015 | 0.074 | ORD | MID|WD40 superfamily; | 0.375 |
| 19 | fp_O | Q9R1A8 | 436 | DYFAIAGVTKKIKVYE | CEEEEECCCCCEEEEE | c1aR-4 | multi-selected | 0.011 | 0.014 | 0.075 | ORD | MID|WD40 superfamily; | 0.286 |
| 20 | fp_O | Q9R1A8 | 436 | DYFAIAGVTKKIKVYE | CEEEEECCCCCEEEEE | c1a-AT-4 | multi | 0.011 | 0.014 | 0.075 | ORD | MID|WD40 superfamily; | 0.286 |
| 21 | fp_O | Q9R1A8 | 436 | DYFAIAGVTKKIKVYE | CEEEEECCCCCEEEEE | c1d-4 | multi | 0.011 | 0.014 | 0.075 | ORD | MID|WD40 superfamily; | 0.286 |
| 22 | fp_O | Q9R1A8 | 519 | WSVDFNLMDPKLLASG | EEEECCCCCCCEEEEE | c1aR-4 | uniq | 0.015 | 0.017 | 0.121 | ORD | MID|WD40 superfamily; | 0.0 |
| 23 | fp_O | Q9R1A8 | 552 | ASIEAKANVCCVKFSP | EEEECCCCEEEEEECC | c1a-AT-4 | multi-selected | 0.01 | 0.017 | 0.068 | ORD | MID|WD40 superfamily; | 0.429 |
| 24 | fp_beta_O | Q9R1A8 | 554 | IEAKANVCCVKFSP | EECCCCEEEEEECC | c2-AT-4 | multi | 0.01 | 0.016 | 0.068 | ORD | MID|WD40 superfamily; | 0.571 |
| 25 | fp_O | Q9R1A8 | 586 | DLRNTKQPIMVFKG | ECCCCCCCEEECCC | c3-AT | uniq | 0.01 | 0.028 | 0.138 | ORD | MID|WD40 superfamily; | 0.286 |
| 26 | fp_beta_O | Q9R1A8 | 612 | GEEIVSASTDSQLKLWN | CCEEEEEECCCCEEEEE | c1c-AT-4 | uniq | 0.021 | 0.01 | 0.155 | ORD | MID|WD40 superfamily; | 0.5 |
| 27 | fp_beta_D | Q9R1A8 | 669 | YYKGLSKTLLTFKFDT | EECCCCCEEEEEEECC | c1a-4 | multi-selected | 0.021 | 0.04 | 0.046 | boundary | MID|WD40 superfamily; | 0.571 |
| 28 | fp_beta_D | Q9R1A8 | 669 | YYKGLSKTLLTFKFDT | EECCCCCEEEEEEECC | c1d-AT-4 | multi | 0.021 | 0.04 | 0.046 | boundary | MID|WD40 superfamily; | 0.571 |
| 29 | fp_D | Q9R1A8 | 678 | LTFKFDTVKSVLDKDR | EEEEECCCCCCCCCCC | c1aR-4 | uniq | 0.179 | 0.25 | 0.179 | boundary | MID|WD40 superfamily; | 0.143 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment