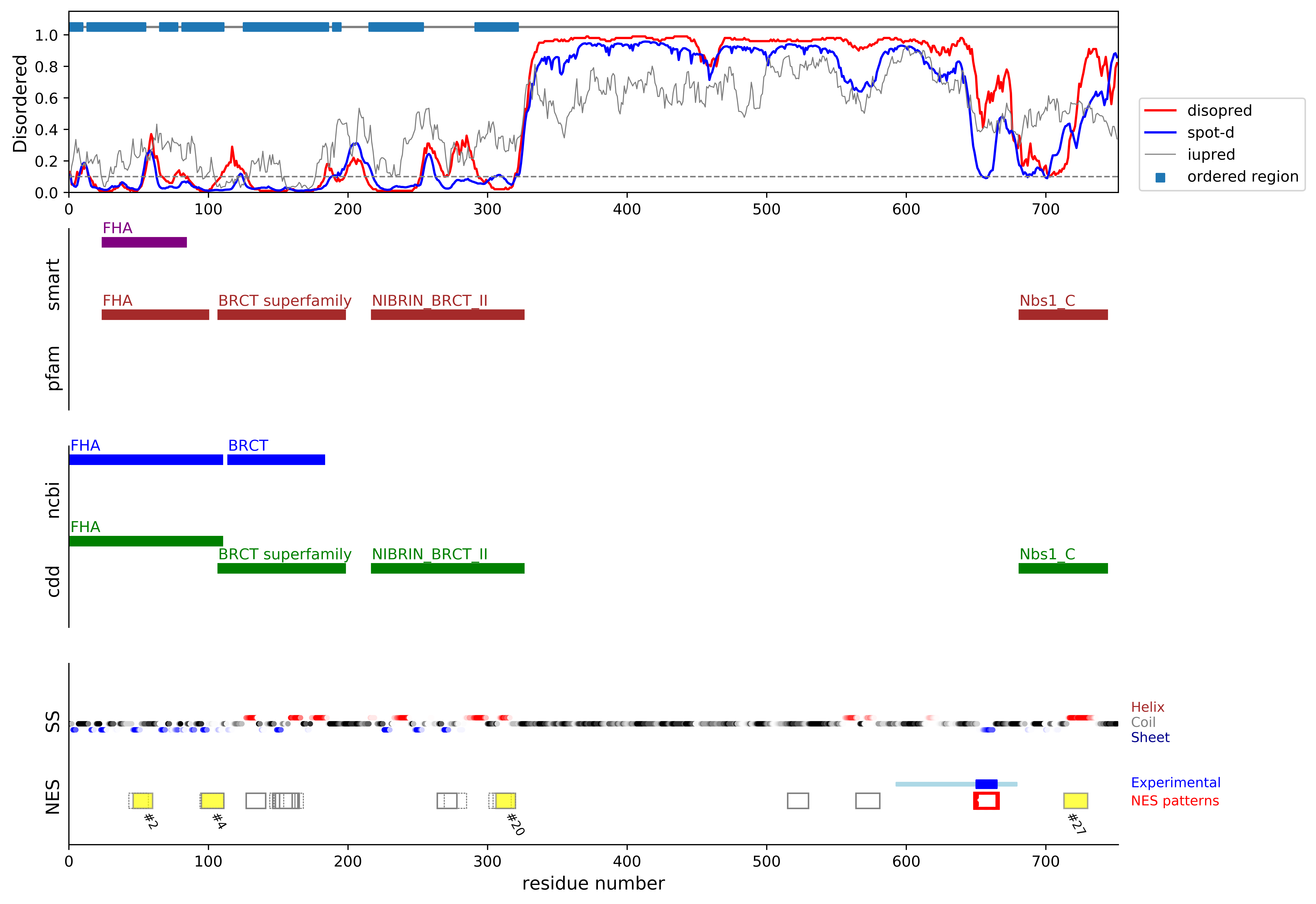

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | Q9R207 | 43 | RNHAVLTVNFPVTS | CCCEEEEEECCCCC | c2-AT-4 | multi | 0.049 | 0.072 | 0.247 | boundary | MID|FHA; | 0.857 |
| 2 | fp_D | Q9R207 | 46 | AVLTVNFPVTSLSQ | EEEEEECCCCCCCC | c3-4 | multi-selected | 0.112 | 0.121 | 0.254 | boundary | MID|FHA; | 0.429 |
| 3 | fp_D | Q9R207 | 94 | GDRVTFGVFESKFRVEY | CCEEEECCCCCCEEEEC | c1c-AT-4 | multi | 0.043 | 0.015 | 0.102 | boundary | boundary|FHA; boundary|BRCT superfamily; | 0.25 |
| 4 | fp_D | Q9R207 | 95 | DRVTFGVFESKFRVEY | CEEEECCCCCCEEEEC | c1aR-4 | multi-selected | 0.045 | 0.014 | 0.098 | boundary | boundary|FHA; boundary|BRCT superfamily; | 0.143 |
| 5 | fp_D | Q9R207 | 95 | DRVTFGVFESKFRVEY | CEEEECCCCCCEEEEC | c1d-4 | multi | 0.045 | 0.014 | 0.098 | boundary | boundary|FHA; boundary|BRCT superfamily; | 0.143 |
| 6 | fp_D | Q9R207 | 127 | VLNQAILQLGGLTA | HHHHHHHHHCCEEE | c3-AT | uniq | 0.037 | 0.028 | 0.169 | boundary | boundary|FHA; boundary|BRCT superfamily; | 0.0 |
| 7 | fp_beta_O | Q9R207 | 144 | TEECTHLVMSAVKVTI | CCCCEEEEECCCCCHH | c1a-AT-4 | multi | 0.012 | 0.021 | 0.129 | ORD | boundary|FHA; MID|BRCT superfamily; | 0.714 |
| 8 | fp_beta_O | Q9R207 | 144 | TEECTHLVMSAVKVTI | CCCCEEEEECCCCCHH | c1d-AT-4 | multi | 0.012 | 0.021 | 0.129 | ORD | boundary|FHA; MID|BRCT superfamily; | 0.714 |
| 9 | fp_O | Q9R207 | 146 | ECTHLVMSAVKVTIKT | CCEEEEECCCCCHHHH | c1a-AT-4 | multi | 0.013 | 0.019 | 0.108 | ORD | MID|BRCT superfamily; | 0.429 |
| 10 | fp_beta_O | Q9R207 | 146 | ECTHLVMSAVKVTI | CCEEEEECCCCCHH | c2-4 | multi-selected | 0.012 | 0.02 | 0.118 | ORD | MID|BRCT superfamily; | 0.571 |
| 11 | fp_O | Q9R207 | 147 | CTHLVMSAVKVTIKT | CEEEEECCCCCHHHH | c1b-AT-4 | multi | 0.013 | 0.019 | 0.099 | ORD | MID|BRCT superfamily; | 0.286 |
| 12 | fp_O | Q9R207 | 148 | THLVMSAVKVTIKTIC | EEEEECCCCCHHHHHH | c1aR-5 | multi-selected | 0.013 | 0.018 | 0.086 | ORD | MID|BRCT superfamily; | 0.143 |
| 13 | fp_O | Q9R207 | 148 | THLVMSAVKVTIKT | EEEEECCCCCHHHH | c2-AT-4 | multi | 0.014 | 0.019 | 0.093 | ORD | MID|BRCT superfamily; | 0.286 |
| 14 | fp_O | Q9R207 | 151 | VMSAVKVTIKTICA | EECCCCCHHHHHHH | c3-4 | multi-selected | 0.014 | 0.019 | 0.063 | ORD | MID|BRCT superfamily; | 0.0 |
| 15 | fp_O | Q9R207 | 154 | AVKVTIKTICALIC | CCCCHHHHHHHHHC | c3-AT | multi | 0.014 | 0.018 | 0.045 | ORD | MID|BRCT superfamily; | 0.0 |
| 16 | fp_beta_D | Q9R207 | 264 | SAPGTCVVDVGITN | CCCCEEEEECCCCC | c2-AT-4 | multi-selected | 0.168 | 0.046 | 0.28 | boundary | MID|NIBRIN_BRCT_II; | 0.714 |
| 17 | fp_D | Q9R207 | 269 | CVVDVGITNTQLIISH | EEEECCCCCCCEEECH | c1d-AT-5 | multi | 0.222 | 0.055 | 0.231 | boundary | MID|NIBRIN_BRCT_II; | 0.0 |
| 18 | fp_D | Q9R207 | 301 | GLRPIPEAEIGLAVIF | CCCCCCCHHHHHHHHH | c1d-AT-5 | multi | 0.027 | 0.089 | 0.343 | boundary | boundary|NIBRIN_BRCT_II; | 0.0 |
| 19 | fp_D | Q9R207 | 304 | PIPEAEIGLAVIFMTT | CCCCHHHHHHHHHCCC | c1a-AT-5 | multi | 0.027 | 0.084 | 0.326 | boundary | boundary|NIBRIN_BRCT_II; | 0.0 |
| 20 | fp_D | Q9R207 | 306 | PEAEIGLAVIFMTT | CCHHHHHHHHHCCC | c2-4 | multi-selected | 0.028 | 0.082 | 0.312 | boundary | boundary|NIBRIN_BRCT_II; | 0.0 |
| 21 | fp_D | Q9R207 | 515 | READTSSVGGMDIEL | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.963 | 0.925 | 0.788 | DISO | 0.0 | |
| 22 | fp_D | Q9R207 | 564 | LLRSTKPELAVQVKVEK | HHCCCCCHHHHHHHCCC | c1c-AT-5 | uniq | 0.929 | 0.684 | 0.607 | DISO | 0.0 | |
| 23 | cand_beta_D | Q9R207 | 649 | PRKLLLTEFRSLVVSN ....**.....*.*.. | CCCEEEEEEEEEEECC | c1a-4 | multi-selected | 0.591 | 0.148 | 0.415 | DISO | boundary|Nbs1_C; | 1.0 |
| 24 | cand_beta_D | Q9R207 | 650 | RKLLLTEFRSLVVSNH ...**.....*.*... | CCEEEEEEEEEEECCC | c1aR-4 | multi-selected | 0.585 | 0.155 | 0.412 | DISO | boundary|Nbs1_C; | 1.0 |
| 25 | cand_beta_D | Q9R207 | 650 | RKLLLTEFRSLVVSN ...**.....*.*.. | CCEEEEEEEEEEECC | c1b-4 | multi | 0.578 | 0.141 | 0.409 | DISO | boundary|Nbs1_C; | 1.0 |
| 26 | cand_beta_D | Q9R207 | 651 | KLLLTEFRSLVVSN ..**.....*.*.. | CEEEEEEEEEEECC | c2-AT-5 | multi | 0.566 | 0.136 | 0.407 | DISO | boundary|Nbs1_C; | 1.0 |
| 27 | fp_D | Q9R207 | 713 | KNTELEEWLKQEMEVQK | CCCHHHHHHHHHHHHHH | c1c-4 | uniq | 0.483 | 0.406 | 0.54 | DISO | boundary|Nbs1_C; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment