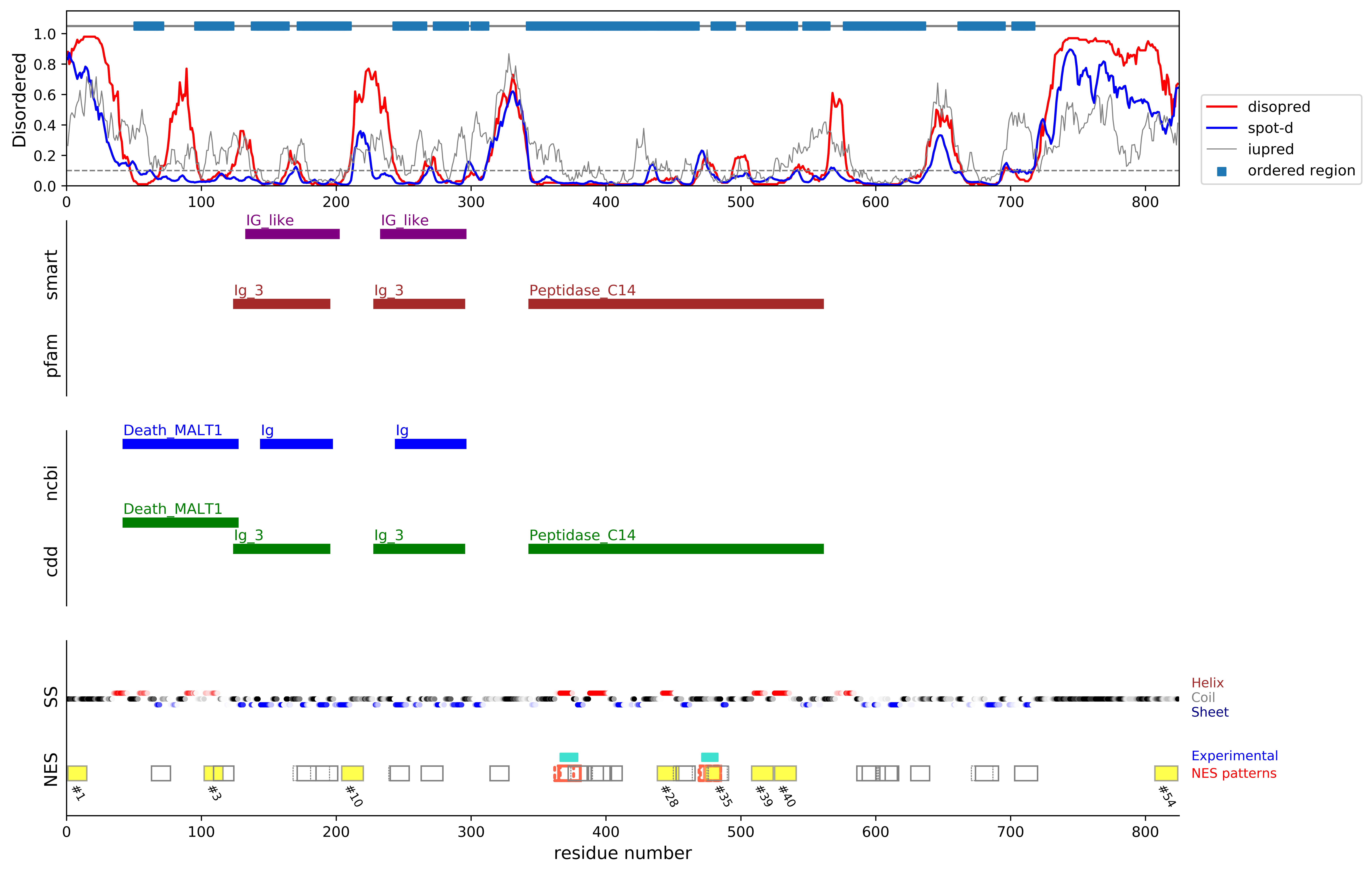

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9UDY8 | 1 | XMSLLGDPLQALPP | CCCCCCCCCCCCCC | c3-4-Nt | uniq | 0.919 | 0.774 | 0.486 | DISO | 0.0 | |
| 2 | fp_beta_D | Q9UDY8 | 63 | SRGRLRLSCLDLEQ | CCCEEEECCCCCCC | c2-4 | uniq | 0.116 | 0.058 | 0.14 | boundary | MID|Death_MALT1; | 0.571 |
| 3 | fp_D | Q9UDY8 | 102 | TVTELSDFLQAMEH | CHHHHHHHHHHCCC | c3-4 | uniq | 0.054 | 0.042 | 0.216 | boundary | boundary|Death_MALT1; boundary|Ig_3; | 0.0 |
| 4 | fp_D | Q9UDY8 | 109 | FLQAMEHTEVLQLLS | HHHHCCCCCCEECCC | c1b-AT-5 | uniq | 0.077 | 0.054 | 0.215 | boundary | boundary|Death_MALT1; boundary|Ig_3; | 0.143 |
| 5 | fp_beta_D | Q9UDY8 | 168 | PNGNTSELIFNAVHVKD | CCCCCCEEEEEEECCCC | c1c-AT-4 | multi | 0.074 | 0.052 | 0.159 | __ | boundary|Ig_3; | 0.75 |
| 6 | fp_beta_D | Q9UDY8 | 171 | NTSELIFNAVHVKD | CCCEEEEEEECCCC | c2-4 | multi-selected | 0.057 | 0.04 | 0.156 | boundary | boundary|Ig_3; | 1.0 |
| 7 | fp_beta_D | Q9UDY8 | 181 | HVKDAGFYVCRVNN | CCCCCCEEEEEEEC | c3-AT | multi | 0.012 | 0.022 | 0.033 | boundary | boundary|Ig_3; | 0.571 |
| 8 | fp_beta_D | Q9UDY8 | 185 | AGFYVCRVNNNFTFEF | CCEEEEEEECCCCCEE | c1aR-4 | multi-selected | 0.011 | 0.023 | 0.033 | boundary | boundary|Ig_3; | 0.714 |
| 9 | fp_beta_D | Q9UDY8 | 185 | AGFYVCRVNNNFTFEF | CCEEEEEEECCCCCEE | c1d-4 | multi | 0.011 | 0.023 | 0.033 | boundary | boundary|Ig_3; | 0.714 |
| 10 | fp_D | Q9UDY8 | 204 | SQLDVCDIPESFQRSV | EEEEEEECCCCCCCCC | c1aR-4 | uniq | 0.306 | 0.14 | 0.161 | boundary | boundary|Ig_3; boundary|Ig_3; | 0.429 |
| 11 | fp_beta_D | Q9UDY8 | 239 | MPGSTLVLQCVAVGS | CCCCCEEEEEEEEEC | c1b-AT-4 | multi | 0.115 | 0.03 | 0.165 | boundary | boundary|Ig_3; | 0.857 |
| 12 | fp_beta_D | Q9UDY8 | 240 | PGSTLVLQCVAVGS | CCCCEEEEEEEEEC | c2-4 | multi-selected | 0.096 | 0.027 | 0.169 | boundary | boundary|Ig_3; | 0.857 |
| 13 | fp_D | Q9UDY8 | 263 | NELPLTHETKKLYMVP | CCCCCCCCCCCEEEEE | c1a-AT-4 | uniq | 0.113 | 0.065 | 0.233 | boundary | boundary|Ig_3; | 0.0 |
| 14 | fp_D | Q9UDY8 | 314 | AVECTEDELNNLGH | CCCCCCCCCCCCCC | c3-AT | uniq | 0.437 | 0.372 | 0.544 | boundary | boundary|Ig_3; boundary|Peptidase_C14; | 0.0 |
| 15 | cand_O | Q9UDY8 | 362 | PLVDVYELTNLLRQ +++++++ | CHHHHHHHHHHHHH | c3-AT | multi | 0.014 | 0.02 | 0.12 | ORD | boundary|Peptidase_C14; | 0.0 |
| 16 | cand_O | Q9UDY8 | 365 | DVYELTNLLRQLDFKV ++++++++ | HHHHHHHHHHHCCCEE | c1a-5 | multi-selected | 0.011 | 0.018 | 0.086 | ORD | MID|Peptidase_C14; | 0.0 |
| 17 | cand_O | Q9UDY8 | 365 | DVYELTNLLRQLDFKV ++++++++ | HHHHHHHHHHHCCCEE | c1d-5 | multi | 0.011 | 0.018 | 0.086 | ORD | MID|Peptidase_C14; | 0.0 |
| 18 | cand_O | Q9UDY8 | 366 | VYELTNLLRQLDFKV ++++++++ | HHHHHHHHHHCCCEE | c1b-AT-4 | multi | 0.011 | 0.018 | 0.076 | ORD | MID|Peptidase_C14; | 0.0 |
| 19 | fp_O | Q9UDY8 | 372 | LLRQLDFKVVSLLD +++++ | HHHHCCCEEEEECC | c2-5 | multi-selected | 0.01 | 0.016 | 0.07 | ORD | MID|Peptidase_C14; | 0.429 |
| 20 | fp_O | Q9UDY8 | 372 | LLRQLDFKVVSLLD +++++ | HHHHCCCEEEEECC | c3-4 | multi-selected | 0.01 | 0.016 | 0.07 | ORD | MID|Peptidase_C14; | 0.429 |
| 21 | fp_beta_O | Q9UDY8 | 372 | LLRQLDFKVVSLLDLTE +++++ | HHHHCCCEEEEECCCCH | c1c-5 | multi-selected | 0.01 | 0.018 | 0.077 | ORD | MID|Peptidase_C14; | 0.625 |
| 22 | fp_beta_O | Q9UDY8 | 374 | RQLDFKVVSLLDLTE +++ | HHCCCEEEEECCCCH | c1b-4 | multi | 0.01 | 0.018 | 0.08 | ORD | MID|Peptidase_C14; | 0.714 |
| 23 | fp_O | Q9UDY8 | 387 | TEYEMRNAVDEFLLLLD | CHHHHHHHHHHHHHHHC | c1c-4 | multi-selected | 0.011 | 0.02 | 0.068 | ORD | MID|Peptidase_C14; | 0.0 |
| 24 | fp_O | Q9UDY8 | 387 | TEYEMRNAVDEFLLLL | CHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.011 | 0.02 | 0.071 | ORD | MID|Peptidase_C14; | 0.0 |
| 25 | fp_O | Q9UDY8 | 387 | TEYEMRNAVDEFLLLL | CHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.011 | 0.02 | 0.071 | ORD | MID|Peptidase_C14; | 0.0 |
| 26 | fp_O | Q9UDY8 | 390 | EMRNAVDEFLLLLD | HHHHHHHHHHHHHC | c3-AT | multi | 0.011 | 0.019 | 0.058 | ORD | MID|Peptidase_C14; | 0.0 |
| 27 | fp_O | Q9UDY8 | 398 | FLLLLDKGVYGLLY | HHHHHCCCCEEEEE | c3-4 | uniq | 0.011 | 0.014 | 0.027 | ORD | MID|Peptidase_C14; | 0.143 |
| 28 | fp_D | Q9UDY8 | 438 | NCLCVQNILKLMQEKE | CCCCHHHHHHHHHHCC | c1aR-4 | uniq | 0.042 | 0.03 | 0.091 | boundary | MID|Peptidase_C14; | 0.0 |
| 29 | fp_beta_D | Q9UDY8 | 450 | QEKETGLNVFLLDMCR | HHCCCCCEEEEEECCC | c1a-AT-4 | multi | 0.022 | 0.025 | 0.089 | boundary | MID|Peptidase_C14; | 0.571 |
| 30 | fp_D | Q9UDY8 | 450 | QEKETGLNVFLLDM | HHCCCCCEEEEEEC | c2-AT-4 | multi | 0.023 | 0.023 | 0.09 | boundary | MID|Peptidase_C14; | 0.429 |
| 31 | fp_beta_D | Q9UDY8 | 452 | KETGLNVFLLDMCR | CCCCCEEEEEECCC | c2-4 | multi-selected | 0.019 | 0.024 | 0.097 | boundary | MID|Peptidase_C14; | 0.714 |
| 32 | cand_D | Q9UDY8 | 469 | DYDDTIPILDALKVTA +++++++ | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi | 0.121 | 0.124 | 0.092 | boundary | MID|Peptidase_C14; | 0.0 |
| 33 | cand_D | Q9UDY8 | 469 | DYDDTIPILDALKVTA +++++++ | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.121 | 0.124 | 0.092 | boundary | MID|Peptidase_C14; | 0.0 |
| 34 | cand_D | Q9UDY8 | 470 | YDDTIPILDALKVTA +++++++ | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.123 | 0.119 | 0.094 | boundary | MID|Peptidase_C14; | 0.0 |
| 35 | cand_D | Q9UDY8 | 473 | TIPILDALKVTA +++++++ | CCCCCCCCCCCC | c2-rev | multi-selected | 0.123 | 0.092 | 0.088 | boundary | MID|Peptidase_C14; | 0.0 |
| 36 | fp_D | Q9UDY8 | 475 | PILDALKVTANIVFGY ++++++ | CCCCCCCCCCCEEEEC | c1d-AT-5 | multi | 0.079 | 0.056 | 0.064 | boundary | MID|Peptidase_C14; | 0.0 |
| 37 | fp_D | Q9UDY8 | 476 | ILDALKVTANIVFG +++++ | CCCCCCCCCCEEEE | c3-AT | multi | 0.076 | 0.053 | 0.053 | boundary | MID|Peptidase_C14; | 0.0 |
| 38 | fp_D | Q9UDY8 | 476 | ILDALKVTANIVFGY +++++ | CCCCCCCCCCEEEEC | c1b-AT-5 | multi | 0.072 | 0.051 | 0.052 | boundary | MID|Peptidase_C14; | 0.143 |
| 39 | fp_D | Q9UDY8 | 508 | NGIFMKFLKDRLLEDK | CHHHHHHHHHHCCCCC | c1aR-4 | uniq | 0.012 | 0.036 | 0.055 | boundary | MID|Peptidase_C14; | 0.0 |
| 40 | fp_D | Q9UDY8 | 525 | ITVLLDEVAEDMGKCH | HHHHHHHHHHHHCCCC | c1aR-4 | uniq | 0.039 | 0.047 | 0.161 | boundary | boundary|Peptidase_C14; | 0.0 |
| 41 | fp_beta_D | Q9UDY8 | 586 | LPESMCLKFDCGVQIQL | CCCCEEEEEEECEEEEE | c1c-4 | multi-selected | 0.013 | 0.019 | 0.057 | boundary | boundary|Peptidase_C14; | 0.875 |
| 42 | fp_beta_D | Q9UDY8 | 590 | MCLKFDCGVQIQLGFAA | EEEEEEECEEEEECCCC | c1cR-4 | multi | 0.011 | 0.013 | 0.042 | boundary | boundary|Peptidase_C14; | 0.875 |
| 43 | fp_beta_D | Q9UDY8 | 590 | MCLKFDCGVQIQLGFAA | EEEEEEECEEEEECCCC | c1c-4 | multi-selected | 0.011 | 0.013 | 0.042 | boundary | boundary|Peptidase_C14; | 0.875 |
| 44 | fp_O | Q9UDY8 | 600 | IQLGFAAEFSNVMIIY | EEECCCCCCCEEEEEE | c1a-4 | multi-selected | 0.01 | 0.008 | 0.03 | ORD | 0.143 | |
| 45 | fp_O | Q9UDY8 | 600 | IQLGFAAEFSNVMIIYT | EEECCCCCCCEEEEEEE | c1cR-4 | multi | 0.01 | 0.007 | 0.031 | ORD | 0.25 | |
| 46 | fp_O | Q9UDY8 | 600 | IQLGFAAEFSNVMIIYT | EEECCCCCCCEEEEEEE | c1c-4 | multi-selected | 0.01 | 0.007 | 0.031 | ORD | 0.25 | |
| 47 | fp_O | Q9UDY8 | 601 | QLGFAAEFSNVMIIY | EECCCCCCCEEEEEE | c1b-AT-5 | multi | 0.01 | 0.007 | 0.03 | ORD | 0.286 | |
| 48 | fp_O | Q9UDY8 | 601 | QLGFAAEFSNVMIIYT | EECCCCCCCEEEEEEE | c1d-AT-5 | multi | 0.01 | 0.007 | 0.031 | ORD | 0.286 | |
| 49 | fp_O | Q9UDY8 | 602 | LGFAAEFSNVMIIY | ECCCCCCCEEEEEE | c2-AT-4 | multi | 0.01 | 0.007 | 0.031 | ORD | 0.286 | |
| 50 | fp_D | Q9UDY8 | 626 | IMCDAYVTDFPLDL | EECCCCCCCCCCCC | c2-AT-5 | uniq | 0.104 | 0.045 | 0.118 | boundary | 0.0 | |
| 51 | fp_D | Q9UDY8 | 671 | RLSSLQKLKEHLVFTV | EECCCCCCCCCEEEEE | c1d-5 | multi | 0.011 | 0.02 | 0.074 | boundary | 0.0 | |
| 52 | fp_beta_D | Q9UDY8 | 674 | SLQKLKEHLVFTVCLSY | CCCCCCCCEEEEEEEEE | c1c-5 | multi-selected | 0.012 | 0.016 | 0.057 | boundary | 0.5 | |
| 53 | fp_beta_D | Q9UDY8 | 703 | QEVNVGKPLIAKLDMHR | EEECCCCCEEEEEECCC | c1c-4 | uniq | 0.063 | 0.104 | 0.373 | boundary | 0.5 | |
| 54 | fp_D | Q9UDY8 | 807 | TTDEIPFSFSDRLRISE | CCCCCCCCCCCCCCCCC | c1c-4 | uniq | 0.692 | 0.449 | 0.397 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment