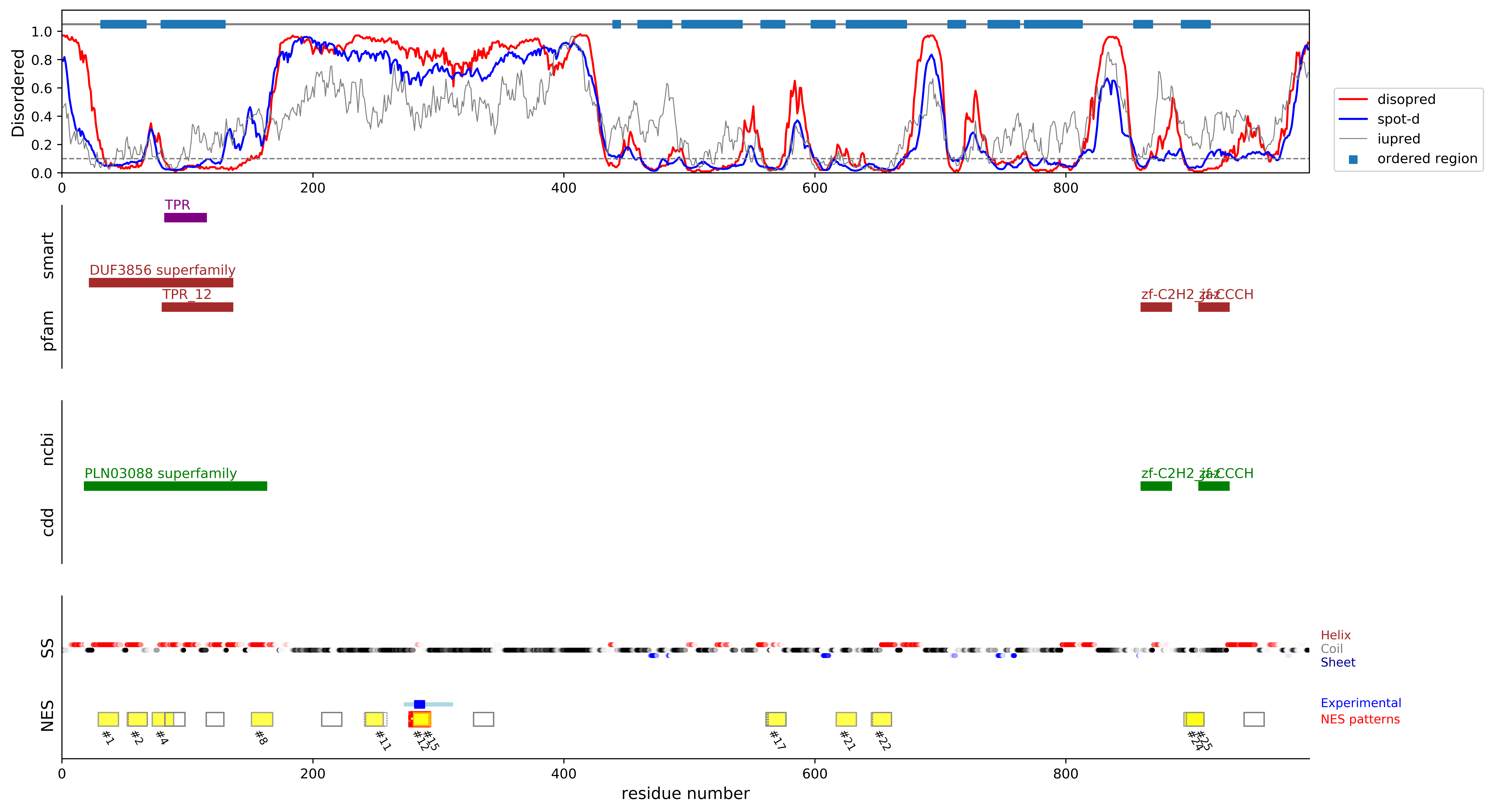

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9UGR2 | 29 | EAFLLKLVQNLFAEGN | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.096 | 0.057 | 0.069 | boundary | boundary|PLN03088 superfamily; | 0.0 |
| 2 | fp_D | Q9UGR2 | 52 | YKQALVQYMEGLNVAD | HHHHHHHHHHHHCCCC | c1a-4 | multi-selected | 0.061 | 0.091 | 0.154 | boundary | MID|PLN03088 superfamily; | 0.0 |
| 3 | fp_D | Q9UGR2 | 53 | KQALVQYMEGLNVAD | HHHHHHHHHHHCCCC | c1b-4 | multi | 0.063 | 0.092 | 0.147 | boundary | MID|PLN03088 superfamily; | 0.0 |
| 4 | fp_D | Q9UGR2 | 72 | DQVALPRELLCKLHVNR | CCCCCHHHHHHHHHHHH | c1c-4 | uniq | 0.148 | 0.089 | 0.098 | boundary | MID|PLN03088 superfamily; | 0.0 |
| 5 | fp_D | Q9UGR2 | 82 | CKLHVNRAACYFTMGL | HHHHHHHHHHHHHCCC | c1a-AT-4 | multi-selected | 0.031 | 0.024 | 0.068 | boundary | MID|PLN03088 superfamily; | 0.0 |
| 6 | fp_D | Q9UGR2 | 82 | CKLHVNRAACYFTMGL | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.031 | 0.024 | 0.068 | boundary | MID|PLN03088 superfamily; | 0.0 |
| 7 | fp_D | Q9UGR2 | 115 | SIRALFRKARALNE | CHHHHHHHHHHHHH | c3-AT | uniq | 0.036 | 0.082 | 0.215 | boundary | MID|PLN03088 superfamily; | 0.0 |
| 8 | fp_D | Q9UGR2 | 151 | SVTQLGQELAQKLGLRV | HHHHHHHHHHHHHHHHH | c1c-5 | uniq | 0.2 | 0.345 | 0.382 | DISO | boundary|PLN03088 superfamily; | 0.0 |
| 9 | fp_D | Q9UGR2 | 207 | ETGNVPDTREQVEIGA | CCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.874 | 0.89 | 0.632 | DISO | 0.0 | |
| 10 | fp_D | Q9UGR2 | 241 | PTMPLFPHVLDLLAPLDS | CCCCCCCCCCCCCCCCCC | c4-4 | multi | 0.942 | 0.838 | 0.408 | DISO | 0.0 | |
| 11 | fp_D | Q9UGR2 | 242 | TMPLFPHVLDLLAP | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.944 | 0.836 | 0.385 | DISO | 0.0 | |
| 12 | cand_D | Q9UGR2 | 277 | FGPELDTLLDSLSLVQ .......*.*...... | CCCCHHHHHHHCCCCC | c1a-4 | multi-selected | 0.843 | 0.669 | 0.409 | DISO | 0.0 | |
| 13 | cand_D | Q9UGR2 | 277 | FGPELDTLLDSLSLVQ .......*.*...... | CCCCHHHHHHHCCCCC | c1d-4 | multi | 0.843 | 0.669 | 0.409 | DISO | 0.0 | |
| 14 | cand_D | Q9UGR2 | 279 | PELDTLLDSLSLVQ .....*.*...... | CCHHHHHHHCCCCC | c2-AT-4 | multi | 0.837 | 0.675 | 0.406 | DISO | 0.0 | |
| 15 | fp_D | Q9UGR2 | 280 | ELDTLLDSLSLVQG ....*.*....... | CHHHHHHHCCCCCC | c3-4 | multi-selected | 0.834 | 0.679 | 0.406 | DISO | 0.0 | |
| 16 | fp_D | Q9UGR2 | 328 | VSSPLPPASFGLVMDP | CCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.844 | 0.691 | 0.464 | DISO | 0.0 | |
| 17 | fp_D | Q9UGR2 | 561 | EHQGIFTFLCEICFDS | HCCCCHHHHHHHHHCC | c1a-4 | multi-selected | 0.051 | 0.034 | 0.02 | boundary | 0.0 | |
| 18 | fp_D | Q9UGR2 | 561 | EHQGIFTFLCEICFDS | HCCCCHHHHHHHHHCC | c1d-4 | multi | 0.051 | 0.034 | 0.02 | boundary | 0.0 | |
| 19 | fp_D | Q9UGR2 | 562 | HQGIFTFLCEICFDS | CCCCHHHHHHHHHCC | c1b-4 | multi | 0.05 | 0.034 | 0.019 | boundary | 0.0 | |
| 20 | fp_D | Q9UGR2 | 563 | QGIFTFLCEICFDS | CCCHHHHHHHHHCC | c2-AT-4 | multi | 0.051 | 0.033 | 0.019 | boundary | 0.0 | |
| 21 | fp_D | Q9UGR2 | 617 | KYSKIRQFQEHFQFDV | CCCCCCCCCCCCEEEC | c1d-4 | uniq | 0.12 | 0.043 | 0.089 | boundary | 0.0 | |
| 22 | fp_D | Q9UGR2 | 645 | DSCHFAHSFIELKVWL | CCCCCCCCHHHHHHHH | c1a-4 | multi-selected | 0.018 | 0.035 | 0.029 | boundary | 0.0 | |
| 23 | fp_D | Q9UGR2 | 646 | SCHFAHSFIELKVWL | CCCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.017 | 0.034 | 0.027 | boundary | 0.0 | |
| 24 | fp_D | Q9UGR2 | 894 | SGWAFRFPMGEFRLCD | CCCCEECCCCCEECCC | c1a-4 | multi-selected | 0.026 | 0.049 | 0.238 | boundary | 0.286 | |
| 25 | fp_D | Q9UGR2 | 896 | WAFRFPMGEFRLCD | CCEECCCCCEECCC | c2-4 | multi-selected | 0.019 | 0.042 | 0.226 | boundary | 0.286 | |
| 26 | fp_D | Q9UGR2 | 942 | LKQKLAKARKDMLLCP | HHHHHHHHHHCCCCCC | c1d-AT-4 | uniq | 0.281 | 0.134 | 0.302 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment