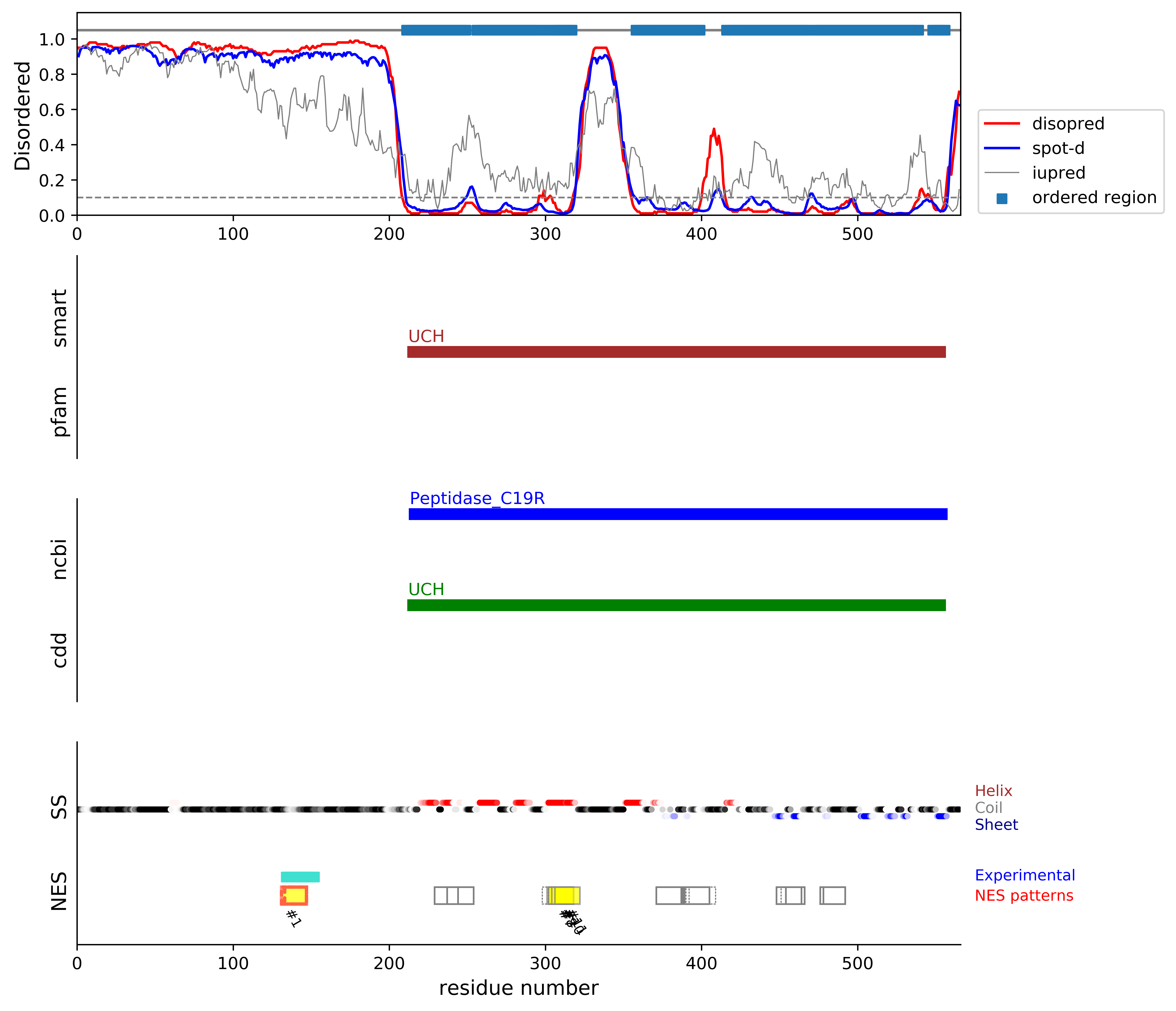

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | Q9UK80 | 131 | GTGELGAALSRLALRP +++++++++++++ | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.934 | 0.897 | 0.608 | DISO | 0.0 | |
| 2 | cand_D | Q9UK80 | 131 | GTGELGAALSRLALRP +++++++++++++ | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.934 | 0.897 | 0.608 | DISO | 0.0 | |
| 3 | cand_D | Q9UK80 | 133 | GELGAALSRLALRP +++++++++++++ | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.935 | 0.898 | 0.615 | DISO | 0.0 | |
| 4 | fp_D | Q9UK80 | 229 | CLSSTRPLRDFCLRR | HHCCCHHHHHHHHCC | c1b-AT-5 | uniq | 0.01 | 0.031 | 0.2 | boundary | boundary|UCH; | 0.0 |
| 5 | fp_D | Q9UK80 | 237 | RDFCLRRDFRQEVPGGG | HHHHHCCHHHHHCCCCC | c1cR-4 | uniq | 0.033 | 0.076 | 0.385 | boundary | MID|UCH; | 0.0 |
| 6 | fp_D | Q9UK80 | 298 | SQQDAQEFLKLLMER | CCCHHHHHHHHHHHH | c1b-AT-4 | multi | 0.067 | 0.02 | 0.169 | boundary | MID|UCH; | 0.0 |
| 7 | fp_D | Q9UK80 | 301 | DAQEFLKLLMERLHLEI | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.052 | 0.016 | 0.187 | boundary | MID|UCH; | 0.0 |
| 8 | fp_D | Q9UK80 | 302 | AQEFLKLLMERLHLEI | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.049 | 0.016 | 0.187 | boundary | MID|UCH; | 0.0 |
| 9 | fp_D | Q9UK80 | 302 | AQEFLKLLMERLHLEI | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.049 | 0.016 | 0.187 | boundary | MID|UCH; | 0.0 |
| 10 | fp_D | Q9UK80 | 304 | EFLKLLMERLHLEI | HHHHHHHHHHHHHH | c2-5 | multi-selected | 0.041 | 0.015 | 0.186 | boundary | MID|UCH; | 0.0 |
| 11 | fp_D | Q9UK80 | 306 | LKLLMERLHLEINRRG | HHHHHHHHHHHHHHCC | c1aR-4 | multi-selected | 0.068 | 0.06 | 0.221 | boundary | MID|UCH; | 0.0 |
| 12 | fp_beta_D | Q9UK80 | 371 | VDLFVGQLKSCLKCQA | HHHHCEEECCEEECCC | c1aR-4 | uniq | 0.014 | 0.036 | 0.067 | boundary | MID|UCH; | 0.571 |
| 13 | fp_D | Q9UK80 | 388 | GYRSTTFEVFCDLSLPI | CCEECCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.074 | 0.039 | 0.076 | boundary | MID|UCH; | 0.0 |
| 14 | fp_D | Q9UK80 | 389 | YRSTTFEVFCDLSLPI | CEECCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.077 | 0.038 | 0.079 | boundary | MID|UCH; | 0.0 |
| 15 | fp_D | Q9UK80 | 389 | YRSTTFEVFCDLSLPI | CEECCCCCCCCCCCCC | c1d-AT-4 | multi | 0.077 | 0.038 | 0.079 | boundary | MID|UCH; | 0.0 |
| 16 | fp_D | Q9UK80 | 390 | RSTTFEVFCDLSLPI | EECCCCCCCCCCCCC | c1b-4 | multi | 0.082 | 0.035 | 0.082 | boundary | MID|UCH; | 0.0 |
| 17 | fp_D | Q9UK80 | 392 | TTFEVFCDLSLPIPKKG | CCCCCCCCCCCCCCCCC | c1cR-4 | multi | 0.172 | 0.042 | 0.104 | boundary | MID|UCH; | 0.0 |
| 18 | fp_beta_O | Q9UK80 | 448 | KKLTVQRFPRILVLHL | EEEEEEECCCEEEEEE | c1aR-4 | multi-selected | 0.014 | 0.013 | 0.147 | ORD | MID|UCH; | 0.571 |
| 19 | fp_beta_O | Q9UK80 | 451 | TVQRFPRILVLHLNR | EEEECCCEEEEEEEC | c1b-5 | multi | 0.011 | 0.011 | 0.119 | ORD | MID|UCH; | 0.571 |
| 20 | fp_beta_O | Q9UK80 | 454 | RFPRILVLHLNR | ECCCEEEEEEEC | c2-rev | multi-selected | 0.01 | 0.009 | 0.099 | ORD | MID|UCH; | 0.833 |
| 21 | fp_O | Q9UK80 | 476 | SSVGVDFPLQRLSLGD | CCCEEECCCCCCCCCC | c1a-4 | multi-selected | 0.022 | 0.04 | 0.19 | ORD | MID|UCH; | 0.286 |
| 22 | fp_O | Q9UK80 | 478 | VGVDFPLQRLSLGD | CEEECCCCCCCCCC | c2-4 | multi-selected | 0.023 | 0.038 | 0.185 | ORD | MID|UCH; | 0.143 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment