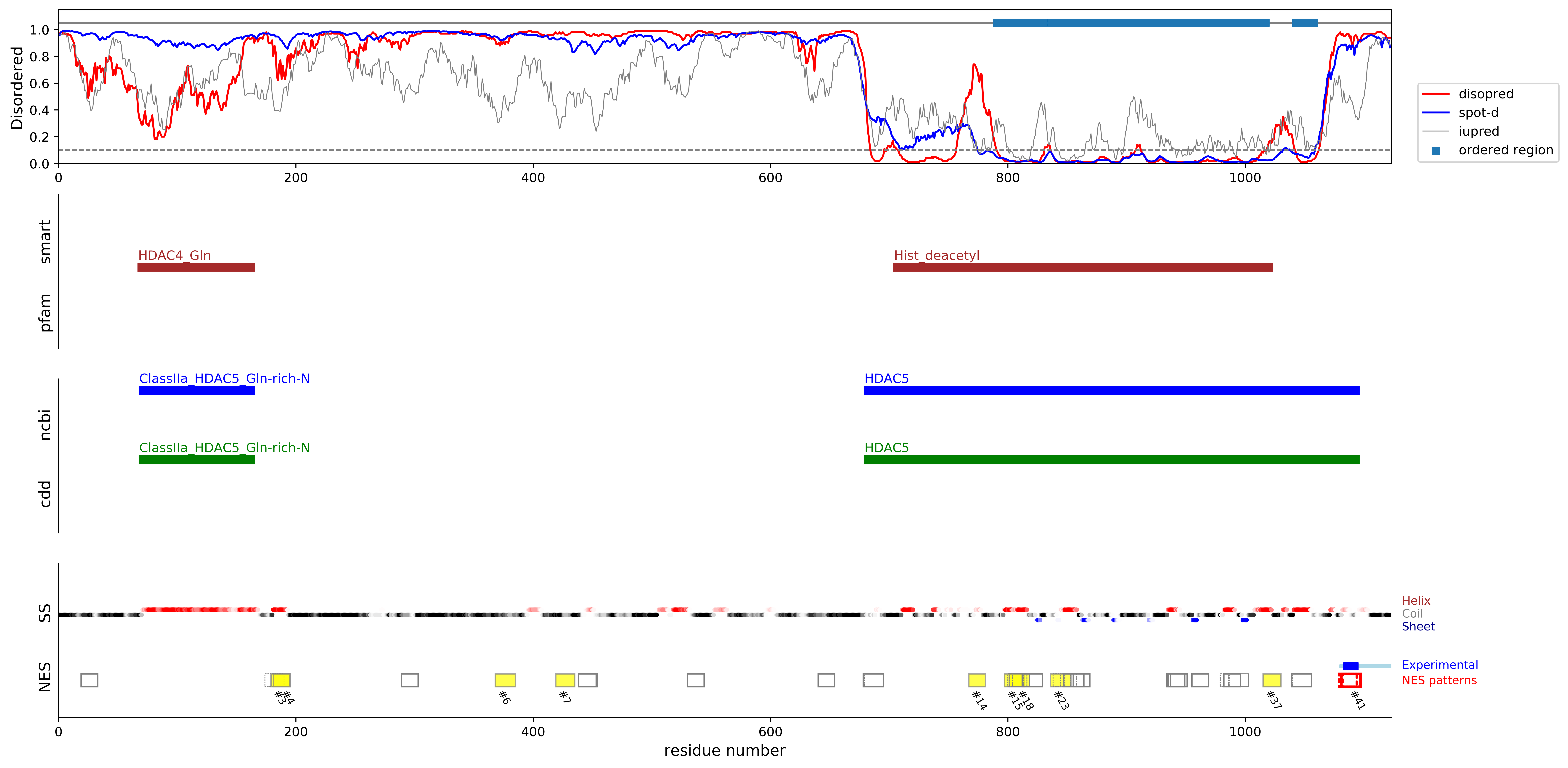

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9UQL6 | 19 | ILPRTSLHSIPVTV | CCCCCCCCCCCCCC | c2-AT-5 | uniq | 0.641 | 0.965 | 0.517 | DISO | 0.0 | |
| 2 | fp_D | Q9UQL6 | 174 | KESAIASTEVKLRLQE | CCCCCCCHHHHHHHHH | c1d-AT-4 | multi | 0.824 | 0.951 | 0.477 | DISO | boundary|ClassIIa_HDAC5_Gln-rich-N; | 0.0 |
| 3 | fp_D | Q9UQL6 | 179 | ASTEVKLRLQEFLLSK | CCHHHHHHHHHHHHHC | c1a-4 | multi-selected | 0.754 | 0.921 | 0.483 | DISO | boundary|ClassIIa_HDAC5_Gln-rich-N; | 0.0 |
| 4 | fp_D | Q9UQL6 | 181 | TEVKLRLQEFLLSK | HHHHHHHHHHHHHC | c2-4 | multi-selected | 0.732 | 0.916 | 0.469 | DISO | boundary|ClassIIa_HDAC5_Gln-rich-N; | 0.0 |
| 5 | fp_D | Q9UQL6 | 289 | VISTFKKRAVEITG | CCCCCCCCCCCCCC | c3-AT | uniq | 0.935 | 0.979 | 0.538 | DISO | 0.0 | |
| 6 | fp_D | Q9UQL6 | 368 | SLPNISLGLQATVTVTN | CCCCCCCCCCCCCCCCC | c1c-5 | uniq | 0.915 | 0.941 | 0.413 | DISO | 0.0 | |
| 7 | fp_D | Q9UQL6 | 419 | STSSIPGCLLGVALEG | CCCCCCCCCCCCCCCC | c1a-4 | uniq | 0.972 | 0.91 | 0.416 | DISO | 0.0 | |
| 8 | fp_D | Q9UQL6 | 438 | PHGHASLLQHVLLLEQ | CCCCCCHHHHHHHHCC | c1d-AT-4 | multi-selected | 0.968 | 0.878 | 0.452 | DISO | 0.0 | |
| 9 | fp_D | Q9UQL6 | 438 | PHGHASLLQHVLLLE | CCCCCCHHHHHHHHC | c1b-AT-4 | multi-selected | 0.968 | 0.882 | 0.466 | DISO | 0.0 | |
| 10 | fp_D | Q9UQL6 | 530 | QLGKILTKTGELPR | HHHCCCCCCCCCCC | c3-AT | uniq | 0.971 | 0.932 | 0.728 | DISO | 0.0 | |
| 11 | fp_D | Q9UQL6 | 640 | QVYQAPLSLATVPH | CCCCCCCCCCCCCC | c3-AT | uniq | 0.956 | 0.926 | 0.589 | DISO | 0.0 | |
| 12 | fp_D | Q9UQL6 | 678 | KHLFTTGVVYDTFMLKH | CCCCCCCCCCCHHHHCC | c1c-AT-4 | multi-selected | 0.198 | 0.374 | 0.298 | DISO | boundary|HDAC5; | 0.0 |
| 13 | fp_D | Q9UQL6 | 679 | HLFTTGVVYDTFMLKH | CCCCCCCCCCHHHHCC | c1d-AT-5 | multi | 0.161 | 0.361 | 0.282 | DISO | boundary|HDAC5; | 0.0 |
| 14 | fp_D | Q9UQL6 | 767 | LLGPISQKMYAVLP | CCCCCHHHHHHCCC | c3-4 | uniq | 0.613 | 0.146 | 0.155 | boundary | MID|HDAC5; | 0.0 |
| 15 | fp_D | Q9UQL6 | 797 | SSSAVRMAVGCLLELAF | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.018 | 0.019 | 0.085 | boundary | MID|HDAC5; | 0.0 |
| 16 | fp_D | Q9UQL6 | 800 | AVRMAVGCLLELAFKV | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.018 | 0.016 | 0.059 | boundary | MID|HDAC5; | 0.0 |
| 17 | fp_D | Q9UQL6 | 801 | VRMAVGCLLELAFKVAA | HHHHHHHHHHHHHHHHC | c1cR-4 | multi | 0.018 | 0.016 | 0.051 | boundary | MID|HDAC5; | 0.0 |
| 18 | fp_D | Q9UQL6 | 801 | VRMAVGCLLELAFKVAA | HHHHHHHHHHHHHHHHC | c1c-4 | multi-selected | 0.018 | 0.016 | 0.051 | boundary | MID|HDAC5; | 0.0 |
| 19 | fp_D | Q9UQL6 | 801 | VRMAVGCLLELAFKV | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.017 | 0.016 | 0.054 | boundary | MID|HDAC5; | 0.0 |
| 20 | fp_D | Q9UQL6 | 804 | AVGCLLELAFKVAA | HHHHHHHHHHHHHC | c3-AT | multi | 0.017 | 0.016 | 0.044 | boundary | MID|HDAC5; | 0.0 |
| 21 | fp_D | Q9UQL6 | 812 | AFKVAAGELKNGFAIIR | HHHHHCCCCCCCEEEEC | c1c-AT-5 | multi-selected | 0.018 | 0.018 | 0.166 | boundary | MID|HDAC5; | 0.0 |
| 22 | fp_D | Q9UQL6 | 813 | FKVAAGELKNGFAIIR | HHHHCCCCCCCEEEEC | c1d-AT-4 | multi | 0.018 | 0.018 | 0.173 | boundary | MID|HDAC5; | 0.0 |
| 23 | fp_D | Q9UQL6 | 836 | ESTAMGFCFFNSVAITA | CCCCCCEEHHHHHHHHH | c1c-4 | multi-selected | 0.025 | 0.024 | 0.098 | __ | MID|HDAC5; | 0.25 |
| 24 | fp_D | Q9UQL6 | 838 | TAMGFCFFNSVAITA | CCCCEEHHHHHHHHH | c1b-4 | multi | 0.017 | 0.017 | 0.082 | __ | MID|HDAC5; | 0.286 |
| 25 | fp_D | Q9UQL6 | 844 | FFNSVAITAKLLQQ | HHHHHHHHHHHHHH | c3-AT | multi | 0.013 | 0.009 | 0.039 | __ | MID|HDAC5; | 0.0 |
| 26 | fp_D | Q9UQL6 | 847 | SVAITAKLLQQKLNVGK | HHHHHHHHHHHHCCCCE | c1c-AT-5 | multi-selected | 0.013 | 0.011 | 0.042 | __ | MID|HDAC5; | 0.0 |
| 27 | fp_D | Q9UQL6 | 848 | VAITAKLLQQKLNVGK | HHHHHHHHHHHCCCCE | c1d-AT-4 | multi | 0.013 | 0.011 | 0.042 | __ | MID|HDAC5; | 0.0 |
| 28 | fp_O | Q9UQL6 | 855 | LQQKLNVGKVLIVD | HHHHCCCCEEEEEE | c2-4 | uniq | 0.013 | 0.01 | 0.079 | ORD | MID|HDAC5; | 0.286 |
| 29 | fp_O | Q9UQL6 | 934 | DVEYLTAFRTVVMPI | CHHHHHHHHHHHCCC | c1b-5 | multi-selected | 0.01 | 0.01 | 0.106 | ORD | MID|HDAC5; | 0.0 |
| 30 | fp_O | Q9UQL6 | 935 | VEYLTAFRTVVMPI | HHHHHHHHHHHCCC | c2-AT-4 | multi-selected | 0.01 | 0.009 | 0.102 | ORD | MID|HDAC5; | 0.0 |
| 31 | fp_O | Q9UQL6 | 937 | YLTAFRTVVMPIAH | HHHHHHHHHCCCCC | c2-AT-5 | multi-selected | 0.01 | 0.008 | 0.088 | ORD | MID|HDAC5; | 0.0 |
| 32 | fp_O | Q9UQL6 | 955 | DVVLVSAGFDAVEG | CEEEECCCCCCCCC | c3-4 | uniq | 0.016 | 0.024 | 0.124 | ORD | MID|HDAC5; | 0.286 |
| 33 | fp_O | Q9UQL6 | 979 | TARCFGHLTRQLMTLAG | CCCHHHHHHHHHHCCCC | c1c-AT-4 | multi | 0.029 | 0.012 | 0.117 | ORD | MID|HDAC5; | 0.0 |
| 34 | fp_O | Q9UQL6 | 982 | CFGHLTRQLMTLAG | HHHHHHHHHHCCCC | c3-4 | multi-selected | 0.031 | 0.011 | 0.113 | ORD | MID|HDAC5; | 0.0 |
| 35 | fp_D | Q9UQL6 | 986 | LTRQLMTLAGGRVVLAL | HHHHHHCCCCCCEEEEC | c1c-AT-4 | multi | 0.026 | 0.011 | 0.139 | boundary | MID|HDAC5; | 0.0 |
| 36 | fp_D | Q9UQL6 | 987 | TRQLMTLAGGRVVLAL | HHHHHCCCCCCEEEEC | c1d-AT-4 | multi | 0.026 | 0.011 | 0.141 | boundary | MID|HDAC5; | 0.0 |
| 37 | fp_D | Q9UQL6 | 1015 | SEACVSALLSVELQP | HHHHHHHHCCCCCCC | c1b-4 | uniq | 0.182 | 0.036 | 0.142 | boundary | MID|HDAC5; | 0.0 |
| 38 | fp_D | Q9UQL6 | 1039 | PNINAVATLEKVIEIQS | CCHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.053 | 0.062 | 0.232 | boundary | MID|HDAC5; | 0.0 |
| 39 | fp_D | Q9UQL6 | 1040 | NINAVATLEKVIEIQS | CHHHHHHHHHHHHHHH | c1d-5 | multi | 0.043 | 0.06 | 0.222 | boundary | MID|HDAC5; | 0.0 |
| 40 | cand_D | Q9UQL6 | 1079 | ETEEAETVSAMALLS .....*....**. | CCCHHHHHHHCCCCC | c1b-AT-4 | multi | 0.95 | 0.872 | 0.485 | DISO | boundary|HDAC5; | 0.0 |
| 41 | cand_D | Q9UQL6 | 1081 | EEAETVSAMALLSVGA .....*....**.... | CHHHHHHHCCCCCCCH | c1a-AT-4 | multi-selected | 0.943 | 0.876 | 0.438 | DISO | boundary|HDAC5; | 0.0 |
| 42 | cand_D | Q9UQL6 | 1082 | EAETVSAMALLSVGA ....*....**.... | HHHHHHHCCCCCCCH | c1b-4 | multi | 0.942 | 0.876 | 0.431 | DISO | boundary|HDAC5; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment