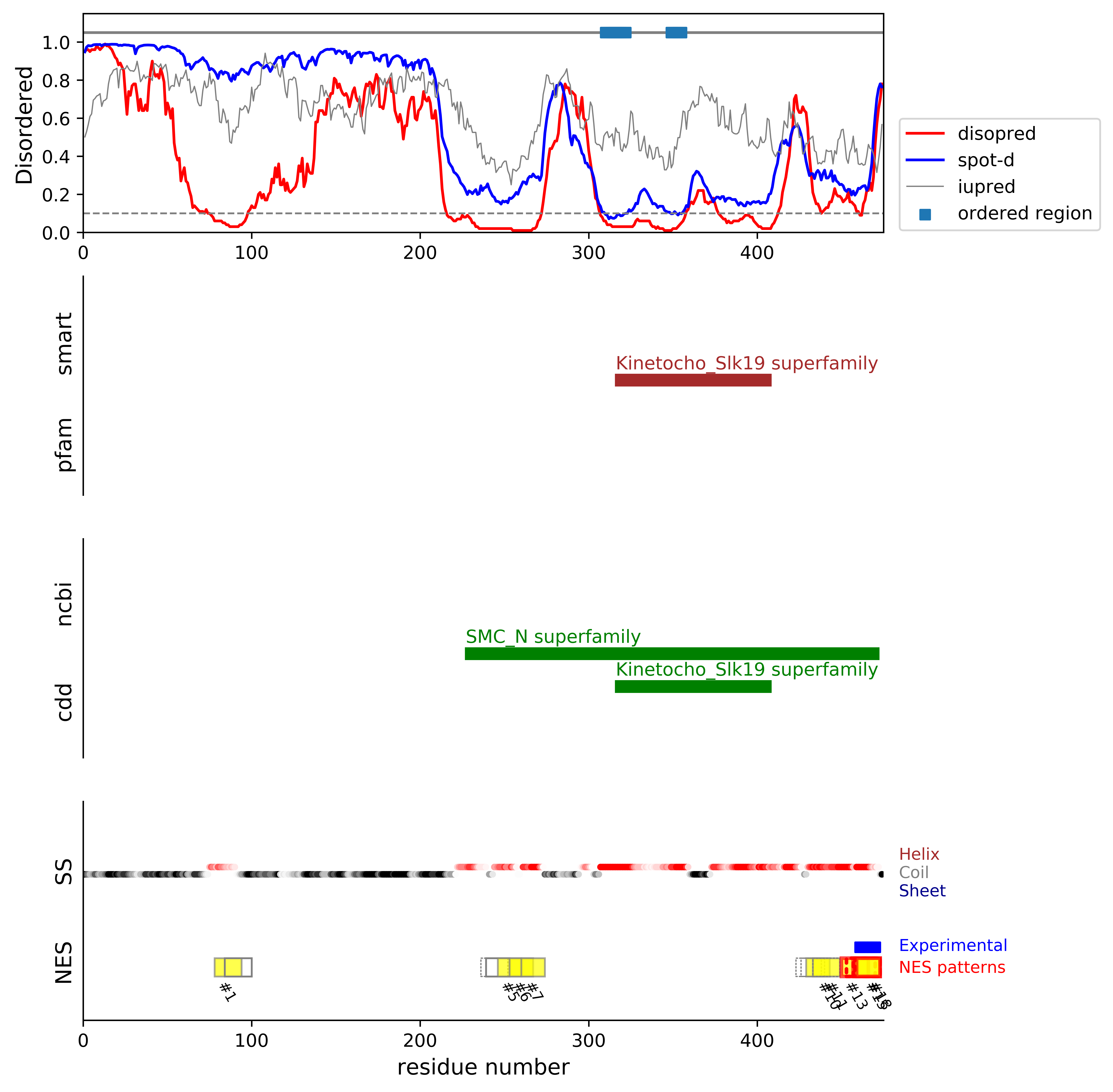

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9URY2 | 78 | LNKRVSDRLQALELNK | HHHHHHHHHHHHHCCC | c1a-4 | uniq | 0.044 | 0.829 | 0.596 | DISO | 0.0 | |
| 2 | fp_D | Q9URY2 | 84 | DRLQALELNKSFDFSG | HHHHHHHCCCCCCCCC | c1d-AT-4 | uniq | 0.051 | 0.834 | 0.592 | DISO | 0.0 | |
| 3 | fp_D | Q9URY2 | 236 | YKAKLAGTYHEISVLQ | HHHHCCCCHHHHHHHH | c1d-AT-4 | multi | 0.02 | 0.195 | 0.372 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 4 | fp_D | Q9URY2 | 239 | KLAGTYHEISVLQN | HCCCCHHHHHHHHH | c3-AT | multi-selected | 0.02 | 0.184 | 0.358 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 5 | fp_D | Q9URY2 | 246 | EISVLQNTIVNVSG | HHHHHHHHHHHHCC | c3-4 | uniq | 0.016 | 0.178 | 0.333 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 6 | fp_D | Q9URY2 | 253 | TIVNVSGQLIAVND | HHHHHCCHHHHHHH | c3-4 | uniq | 0.011 | 0.234 | 0.365 | DISO | MID|SMC_N superfamily; | 0.0 |
| 7 | fp_D | Q9URY2 | 260 | QLIAVNDQLQQLRS | HHHHHHHHHHHHHH | c3-4 | uniq | 0.038 | 0.29 | 0.494 | DISO | MID|SMC_N superfamily; | 0.0 |
| 8 | fp_D | Q9URY2 | 423 | QSQQTGDLETLRLQL | HHHHCCCHHHHHHHH | c1b-AT-4 | multi | 0.443 | 0.396 | 0.464 | DISO | MID|SMC_N superfamily; boundary|Kinetocho_Slk19 superfamily; | 0.0 |
| 9 | fp_D | Q9URY2 | 426 | QTGDLETLRLQLQA | HCCCHHHHHHHHHH | c2-AT-4 | multi | 0.346 | 0.347 | 0.433 | DISO | MID|SMC_N superfamily; boundary|Kinetocho_Slk19 superfamily; | 0.0 |
| 10 | fp_D | Q9URY2 | 429 | DLETLRLQLQALQE | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.236 | 0.31 | 0.426 | DISO | MID|SMC_N superfamily; boundary|Kinetocho_Slk19 superfamily; | 0.0 |
| 11 | fp_D | Q9URY2 | 433 | LRLQLQALQEELRVER | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.159 | 0.296 | 0.412 | DISO | MID|SMC_N superfamily; boundary|Kinetocho_Slk19 superfamily; | 0.0 |
| 12 | fp_D | Q9URY2 | 433 | LRLQLQALQEELRVER | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.159 | 0.296 | 0.412 | DISO | MID|SMC_N superfamily; boundary|Kinetocho_Slk19 superfamily; | 0.0 |
| 13 | cand_D | Q9URY2 | 450 | ERQQLIQMSEDLVIAM *+*++ | HHHHHHHHHHHHHHHH | c1d-4 | multi-selected | 0.153 | 0.226 | 0.421 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 14 | cand_D | Q9URY2 | 453 | QLIQMSEDLVIAMDQLN *+*++++*+ | HHHHHHHHHHHHHHHHH | c1cR-5 | multi | 0.172 | 0.253 | 0.407 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 15 | cand_D | Q9URY2 | 457 | MSEDLVIAMDQLNLEQ *+*++++*+* | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.244 | 0.344 | 0.399 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 16 | cand_D | Q9URY2 | 457 | MSEDLVIAMDQLNLEQ *+*++++*+* | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.244 | 0.344 | 0.399 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 17 | cand_D | Q9URY2 | 458 | SEDLVIAMDQLNLEQ *+*++++*+* | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.25 | 0.352 | 0.391 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 18 | cand_D | Q9URY2 | 459 | EDLVIAMDQLNLEQ *+*++++*+* | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.258 | 0.364 | 0.389 | DISO | boundary|SMC_N superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment