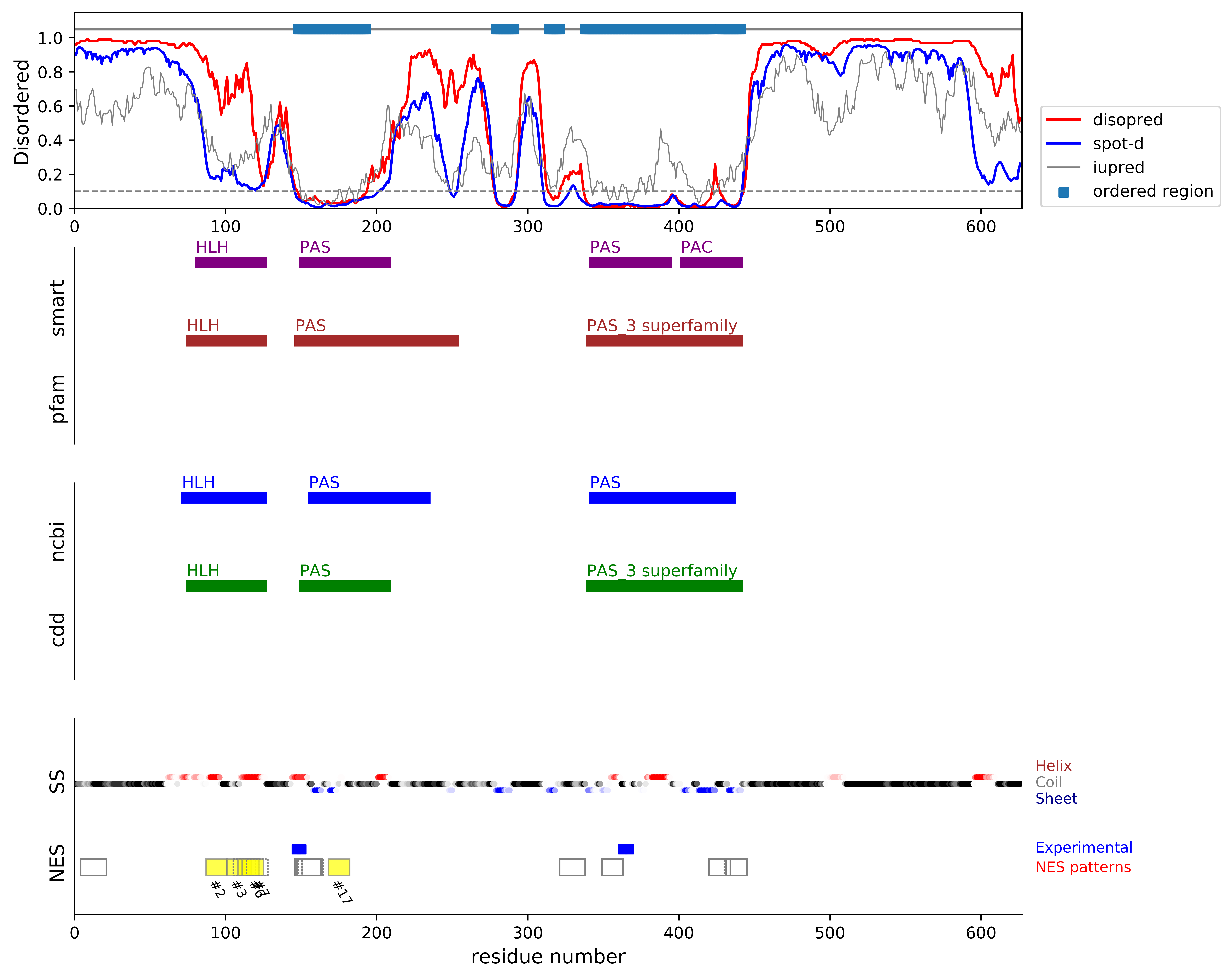

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9WTL8 | 4 | QRMDISSTISDFMSPGP | CCCCCCCCCCCCCCCCC | c1cR-4 | uniq | 0.984 | 0.899 | 0.578 | DISO | 0.0 | |
| 2 | fp_D | Q9WTL8 | 87 | KMNSFIDELASLVP | CHHHHHHHHHCCCC | c3-4 | uniq | 0.719 | 0.202 | 0.322 | DISO | boundary|HLH; | 0.0 |
| 3 | fp_D | Q9WTL8 | 101 | TCNAMSRKLDKLTVLR | CCCCCCCCCHHHHHHH | c1a-4 | multi-selected | 0.726 | 0.17 | 0.274 | DISO | boundary|HLH; | 0.0 |
| 4 | fp_D | Q9WTL8 | 105 | MSRKLDKLTVLRMAV | CCCCCHHHHHHHHHH | c1b-4 | multi | 0.699 | 0.147 | 0.282 | DISO | boundary|HLH; | 0.0 |
| 5 | fp_D | Q9WTL8 | 105 | MSRKLDKLTVLRMAVQH | CCCCCHHHHHHHHHHHH | c1c-AT-4 | multi | 0.663 | 0.143 | 0.295 | DISO | boundary|HLH; | 0.0 |
| 6 | fp_D | Q9WTL8 | 108 | KLDKLTVLRMAVQH | CCHHHHHHHHHHHH | c2-5 | multi-selected | 0.656 | 0.128 | 0.308 | DISO | boundary|HLH; | 0.0 |
| 7 | fp_D | Q9WTL8 | 111 | KLTVLRMAVQHMKT | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.545 | 0.125 | 0.333 | DISO | boundary|HLH; | 0.0 |
| 8 | fp_D | Q9WTL8 | 114 | VLRMAVQHMKTLRG | HHHHHHHHHHHHHC | c3-AT | multi | 0.418 | 0.133 | 0.372 | DISO | boundary|HLH; | 0.0 |
| 9 | fp_D | Q9WTL8 | 146 | ELKHLILRAADGFLFVV * * | HHHHHHHHHCCCEEEEE | c1c-AT-5 | multi-selected | 0.061 | 0.032 | 0.085 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 10 | fp_D | Q9WTL8 | 147 | LKHLILRAADGFLFVVG * * | HHHHHHHHCCCEEEEEE | c1c-AT-4 | multi-selected | 0.056 | 0.027 | 0.073 | boundary | boundary|HLH; boundary|PAS; | 0.125 |
| 11 | fp_D | Q9WTL8 | 147 | LKHLILRAADGFLFVV * * | HHHHHHHHCCCEEEEE | c1a-AT-4 | multi-selected | 0.057 | 0.028 | 0.075 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 12 | fp_D | Q9WTL8 | 147 | LKHLILRAADGFLFVV * * | HHHHHHHHCCCEEEEE | c1d-AT-4 | multi | 0.057 | 0.028 | 0.075 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 13 | fp_D | Q9WTL8 | 148 | KHLILRAADGFLFVVG * | HHHHHHHCCCEEEEEE | c1d-AT-4 | multi | 0.053 | 0.024 | 0.064 | boundary | boundary|HLH; boundary|PAS; | 0.143 |
| 14 | fp_D | Q9WTL8 | 148 | KHLILRAADGFLFVV * | HHHHHHHCCCEEEEE | c1b-AT-4 | multi | 0.053 | 0.026 | 0.065 | boundary | boundary|HLH; boundary|PAS; | 0.143 |
| 15 | fp_D | Q9WTL8 | 150 | LILRAADGFLFVVG * | HHHHHCCCEEEEEE | c3-AT | multi | 0.049 | 0.021 | 0.048 | boundary | boundary|HLH; boundary|PAS; | 0.286 |
| 16 | fp_D | Q9WTL8 | 151 | ILRAADGFLFVVGC | HHHHCCCEEEEEEC | c3-AT | multi | 0.048 | 0.019 | 0.046 | boundary | boundary|HLH; boundary|PAS; | 0.429 |
| 17 | fp_D | Q9WTL8 | 168 | KILFVSESVFKILN | CEEEEECCHHCCCC | c3-4 | uniq | 0.034 | 0.019 | 0.058 | boundary | boundary|PAS; | 0.429 |
| 18 | fp_D | Q9WTL8 | 321 | HSHMVPQPANGEIRVKS | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.18 | 0.073 | 0.374 | boundary | boundary|PAS_3 superfamily; | 0.0 |
| 19 | fp_D | Q9WTL8 | 349 | KFVFVDQRATAILA | EEEEECHHHHHHHC | c3-AT | uniq | 0.01 | 0.024 | 0.093 | boundary | boundary|PAS_3 superfamily; | 0.286 |
| 20 | fp_D | Q9WTL8 | 420 | WFSFMNPWTKEVEY | EEEEECCCCCCCCE | c3-AT | uniq | 0.092 | 0.023 | 0.137 | boundary | boundary|PAS_3 superfamily; | 0.286 |
| 21 | fp_beta_D | Q9WTL8 | 430 | EVEYIVSTNTVVLAN | CCCEEEEEEEEECCC | c1b-AT-5 | multi | 0.047 | 0.061 | 0.221 | boundary | boundary|PAS_3 superfamily; | 1.0 |
| 22 | fp_beta_D | Q9WTL8 | 431 | VEYIVSTNTVVLAN | CCEEEEEEEEECCC | c2-AT-4 | multi-selected | 0.046 | 0.063 | 0.228 | boundary | boundary|PAS_3 superfamily; | 1.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment