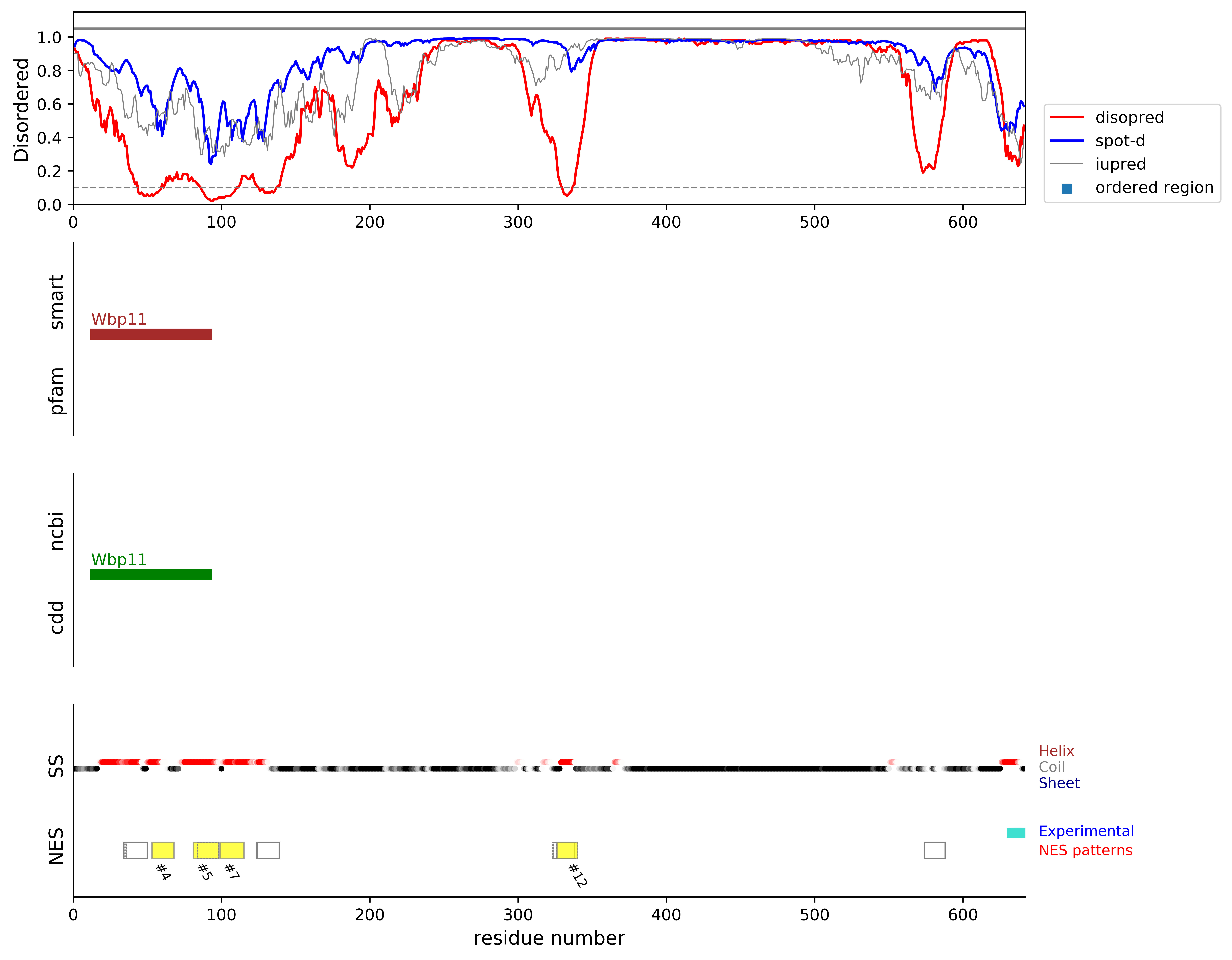

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9Y2W2 | 34 | KKQRMMVRAAVLKMKD | HHHHHHHHHHHHHCCC | c1a-AT-4 | multi-selected | 0.168 | 0.766 | 0.522 | DISO | MID|Wbp11; | 0.0 |
| 2 | fp_D | Q9Y2W2 | 35 | KQRMMVRAAVLKMKD | HHHHHHHHHHHHCCC | c1b-AT-4 | multi | 0.152 | 0.761 | 0.518 | DISO | MID|Wbp11; | 0.0 |
| 3 | fp_D | Q9Y2W2 | 36 | QRMMVRAAVLKMKD | HHHHHHHHHHHCCC | c2-AT-4 | multi | 0.138 | 0.754 | 0.518 | DISO | MID|Wbp11; | 0.0 |
| 4 | fp_D | Q9Y2W2 | 53 | IIRDMEKLDEMEFNP | HHHHHHHHHHCCCCC | c1b-5 | uniq | 0.105 | 0.557 | 0.547 | DISO | MID|Wbp11; | 0.0 |
| 5 | fp_D | Q9Y2W2 | 81 | KRKKLRETFERILRLYE | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.069 | 0.477 | 0.394 | DISO | boundary|Wbp11; | 0.0 |
| 6 | fp_D | Q9Y2W2 | 84 | KLRETFERILRLYE | HHHHHHHHHHHHHH | c3-AT | multi | 0.054 | 0.424 | 0.376 | DISO | boundary|Wbp11; | 0.0 |
| 7 | fp_D | Q9Y2W2 | 99 | ENPDIYKELRKLEVEY | CCHHHHHHHHHHHHHH | c1a-4 | uniq | 0.074 | 0.499 | 0.396 | DISO | boundary|Wbp11; | 0.0 |
| 8 | fp_D | Q9Y2W2 | 124 | YFDAVKNAQHVEVES | HHHHHHHHCCCCCCC | c1b-AT-5 | uniq | 0.084 | 0.569 | 0.458 | DISO | boundary|Wbp11; | 0.0 |
| 9 | fp_D | Q9Y2W2 | 323 | NMKELTPLQAMMLRM | CCCCCCHHHHHHHHH | c1b-5 | multi | 0.16 | 0.918 | 0.876 | DISO | 0.0 | |
| 10 | fp_D | Q9Y2W2 | 324 | MKELTPLQAMMLRM | CCCCCHHHHHHHHH | c2-AT-4 | multi | 0.143 | 0.914 | 0.876 | DISO | 0.0 | |
| 11 | fp_D | Q9Y2W2 | 326 | ELTPLQAMMLRMAG | CCCHHHHHHHHHHC | c2-AT-5 | multi | 0.114 | 0.894 | 0.891 | DISO | 0.0 | |
| 12 | fp_D | Q9Y2W2 | 326 | ELTPLQAMMLRMAG | CCCHHHHHHHHHHC | c3-4 | multi-selected | 0.114 | 0.894 | 0.891 | DISO | 0.0 | |
| 13 | fp_D | Q9Y2W2 | 574 | EITRFVPTALRVRR | CCCCCCCCCCCCCC | c3-AT | uniq | 0.301 | 0.783 | 0.682 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment