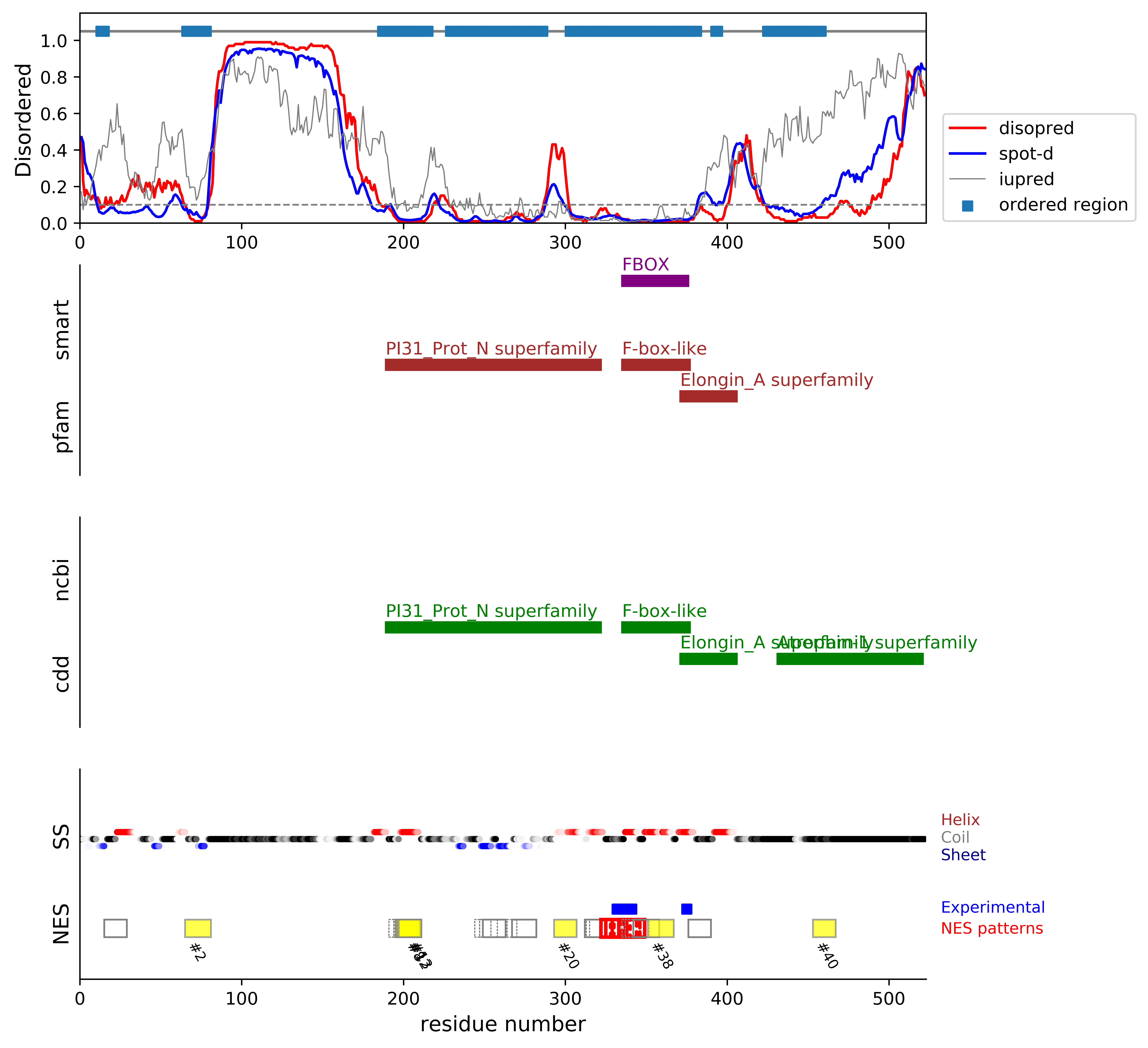

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9Y3I1 | 15 | EVPETEPTLGHLRS | ECCCCCCCHHHHHH | c3-AT | uniq | 0.116 | 0.066 | 0.485 | DISO | 0.0 | |
| 2 | fp_D | Q9Y3I1 | 65 | SYGIVSGDLICLILQD | HCCCCCCCEEEEECCC | c1a-4 | uniq | 0.044 | 0.065 | 0.239 | boundary | 0.429 | |
| 3 | fp_D | Q9Y3I1 | 191 | DCSDANDALIVLIHLLM | CCCCCCCHHHHHHHHHH | c1c-AT-4 | multi | 0.024 | 0.038 | 0.118 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 4 | fp_D | Q9Y3I1 | 194 | DANDALIVLIHLLMLES | CCCCHHHHHHHHHHHHC | c1c-AT-4 | multi | 0.014 | 0.025 | 0.1 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 5 | fp_D | Q9Y3I1 | 194 | DANDALIVLIHLLMLE | CCCCHHHHHHHHHHHH | c1a-AT-4 | multi | 0.014 | 0.025 | 0.098 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 6 | fp_D | Q9Y3I1 | 194 | DANDALIVLIHLLMLE | CCCCHHHHHHHHHHHH | c1d-AT-4 | multi | 0.014 | 0.025 | 0.098 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 7 | fp_D | Q9Y3I1 | 194 | DANDALIVLIHLLM | CCCCHHHHHHHHHH | c2-AT-4 | multi | 0.014 | 0.026 | 0.1 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 8 | fp_D | Q9Y3I1 | 195 | ANDALIVLIHLLMLES | CCCHHHHHHHHHHHHC | c1a-4 | multi-selected | 0.012 | 0.022 | 0.094 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 9 | fp_D | Q9Y3I1 | 195 | ANDALIVLIHLLMLES | CCCHHHHHHHHHHHHC | c1d-4 | multi | 0.012 | 0.022 | 0.094 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 10 | fp_D | Q9Y3I1 | 195 | ANDALIVLIHLLMLE | CCCHHHHHHHHHHHH | c1b-4 | multi | 0.012 | 0.022 | 0.092 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 11 | fp_D | Q9Y3I1 | 196 | NDALIVLIHLLMLES | CCHHHHHHHHHHHHC | c1b-4 | multi | 0.011 | 0.02 | 0.092 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 12 | fp_D | Q9Y3I1 | 196 | NDALIVLIHLLMLE | CCHHHHHHHHHHHH | c2-4 | multi-selected | 0.011 | 0.02 | 0.09 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 13 | fp_D | Q9Y3I1 | 197 | DALIVLIHLLMLES | CHHHHHHHHHHHHC | c2-4 | multi-selected | 0.01 | 0.019 | 0.093 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 14 | fp_beta_O | Q9Y3I1 | 244 | EGSSATLTCVPLGN | CCCCEEEEEEECCC | c2-AT-4 | multi | 0.01 | 0.016 | 0.076 | ORD | MID|PI31_Prot_N superfamily; | 0.857 |
| 15 | fp_beta_O | Q9Y3I1 | 247 | SATLTCVPLGNLIVVNA | CEEEEEEECCCEEEEEE | c1c-AT-4 | multi | 0.011 | 0.01 | 0.059 | ORD | MID|PI31_Prot_N superfamily; | 0.625 |
| 16 | fp_beta_O | Q9Y3I1 | 247 | SATLTCVPLGNLIVVN | CEEEEEEECCCEEEEE | c1a-AT-4 | multi | 0.01 | 0.01 | 0.059 | ORD | MID|PI31_Prot_N superfamily; | 0.571 |
| 17 | fp_beta_O | Q9Y3I1 | 249 | TLTCVPLGNLIVVN | EEEEEECCCEEEEE | c2-5 | multi-selected | 0.01 | 0.009 | 0.049 | ORD | MID|PI31_Prot_N superfamily; | 0.571 |
| 18 | fp_beta_O | Q9Y3I1 | 254 | PLGNLIVVNATLKINN | ECCCEEEEEEEEEECC | c1d-5 | multi | 0.021 | 0.012 | 0.048 | ORD | MID|PI31_Prot_N superfamily; | 1.0 |
| 19 | fp_beta_D | Q9Y3I1 | 267 | INNEIRSVKRLQLLP | ECCCCCCEEEEECCC | c1b-4 | uniq | 0.033 | 0.019 | 0.06 | boundary | MID|PI31_Prot_N superfamily; | 0.571 |
| 20 | fp_D | Q9Y3I1 | 293 | NVANIYKDLQKLSR | CCHHHHHHHHHHHH | c3-4 | uniq | 0.249 | 0.11 | 0.058 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 21 | fp_D | Q9Y3I1 | 312 | LVYPLLAFTRQALNLPD | CCHHHHHHHHHHCCCCC | c1c-AT-5 | multi-selected | 0.046 | 0.029 | 0.021 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 22 | fp_D | Q9Y3I1 | 313 | VYPLLAFTRQALNLPD | CHHHHHHHHHHCCCCC | c1d-AT-4 | multi | 0.048 | 0.029 | 0.021 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 23 | fp_D | Q9Y3I1 | 315 | PLLAFTRQALNLPD | HHHHHHHHHCCCCC | c3-AT | multi | 0.052 | 0.029 | 0.021 | boundary | boundary|PI31_Prot_N superfamily; | 0.0 |
| 24 | cand_O | Q9Y3I1 | 322 | QALNLPDVFGLVVLPLEL *++*+*+* | HHCCCCCCCCCCCCCHHH | c4-4 | multi | 0.044 | 0.03 | 0.014 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 25 | cand_O | Q9Y3I1 | 322 | QALNLPDVFGLVVLPL *++*+* | HHCCCCCCCCCCCCCH | c1a-4 | multi-selected | 0.048 | 0.032 | 0.015 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 26 | cand_O | Q9Y3I1 | 322 | QALNLPDVFGLVVLPL *++*+* | HHCCCCCCCCCCCCCH | c1d-4 | multi | 0.048 | 0.032 | 0.015 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 27 | cand_O | Q9Y3I1 | 322 | QALNLPDVFGLVVLP *++*+ | HHCCCCCCCCCCCCC | c1b-4 | multi | 0.049 | 0.033 | 0.016 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 28 | cand_O | Q9Y3I1 | 325 | NLPDVFGLVVLP *++*+ | CCCCCCCCCCCC | c2-rev | multi-selected | 0.043 | 0.033 | 0.017 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 29 | cand_O | Q9Y3I1 | 328 | DVFGLVVLPLELKLRI *++*+*+*+* | CCCCCCCCCHHHHHHH | c1aR-4 | multi | 0.026 | 0.024 | 0.014 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 30 | cand_O | Q9Y3I1 | 329 | VFGLVVLPLELKLRIFR *++*+*+*+* | CCCCCCCCHHHHHHHHC | c1c-5 | multi | 0.024 | 0.022 | 0.013 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 31 | cand_O | Q9Y3I1 | 330 | FGLVVLPLELKLRIFR *++*+*+*+* | CCCCCCCHHHHHHHHC | c1d-4 | multi | 0.023 | 0.02 | 0.013 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 32 | cand_O | Q9Y3I1 | 333 | VVLPLELKLRIFRLLD ++*+*+*+* | CCCCHHHHHHHHCCCC | c1a-5 | multi-selected | 0.016 | 0.017 | 0.01 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 33 | cand_O | Q9Y3I1 | 335 | LPLELKLRIFRLLD *+*+*+* | CCHHHHHHHHCCCC | c2-4 | multi | 0.014 | 0.015 | 0.01 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 34 | fp_O | Q9Y3I1 | 335 | LPLELKLRIFRLLDVRS *+*+*+* | CCHHHHHHHHCCCCHHH | c1c-4 | multi | 0.013 | 0.015 | 0.011 | ORD | boundary|PI31_Prot_N superfamily; | 0.0 |
| 35 | fp_O | Q9Y3I1 | 337 | LELKLRIFRLLDVRS *+*+* | HHHHHHHHCCCCHHH | c1b-4 | multi | 0.012 | 0.015 | 0.011 | ORD | boundary|PI31_Prot_N superfamily; small|F-box-like; | 0.0 |
| 36 | fp_O | Q9Y3I1 | 342 | RIFRLLDVRSVLSLSA | HHHCCCCHHHHHHHHH | c1aR-4 | multi-selected | 0.01 | 0.013 | 0.016 | ORD | boundary|PI31_Prot_N superfamily; small|F-box-like; | 0.0 |
| 37 | fp_O | Q9Y3I1 | 342 | RIFRLLDVRSVLSLSA | HHHCCCCHHHHHHHHH | c1d-5 | multi | 0.01 | 0.013 | 0.016 | ORD | boundary|PI31_Prot_N superfamily; small|F-box-like; | 0.0 |
| 38 | fp_D | Q9Y3I1 | 351 | SVLSLSAVCRDLFTAS | HHHHHHHHHHHHHHHC | c1aR-4 | uniq | 0.01 | 0.014 | 0.037 | boundary | boundary|PI31_Prot_N superfamily; small|F-box-like; | 0.0 |
| 39 | fp_D | Q9Y3I1 | 376 | YLRDFRDNTVRVQD | HHHHCCCCCCCCCC | c3-AT | uniq | 0.051 | 0.105 | 0.121 | boundary | small|Elongin_A superfamily; | 0.0 |
| 40 | fp_D | Q9Y3I1 | 453 | TLPYVGDPISSLIP | CCCCCCCCCCCCCC | c3-4 | uniq | 0.038 | 0.11 | 0.539 | boundary | MID|Atrophin-1 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment