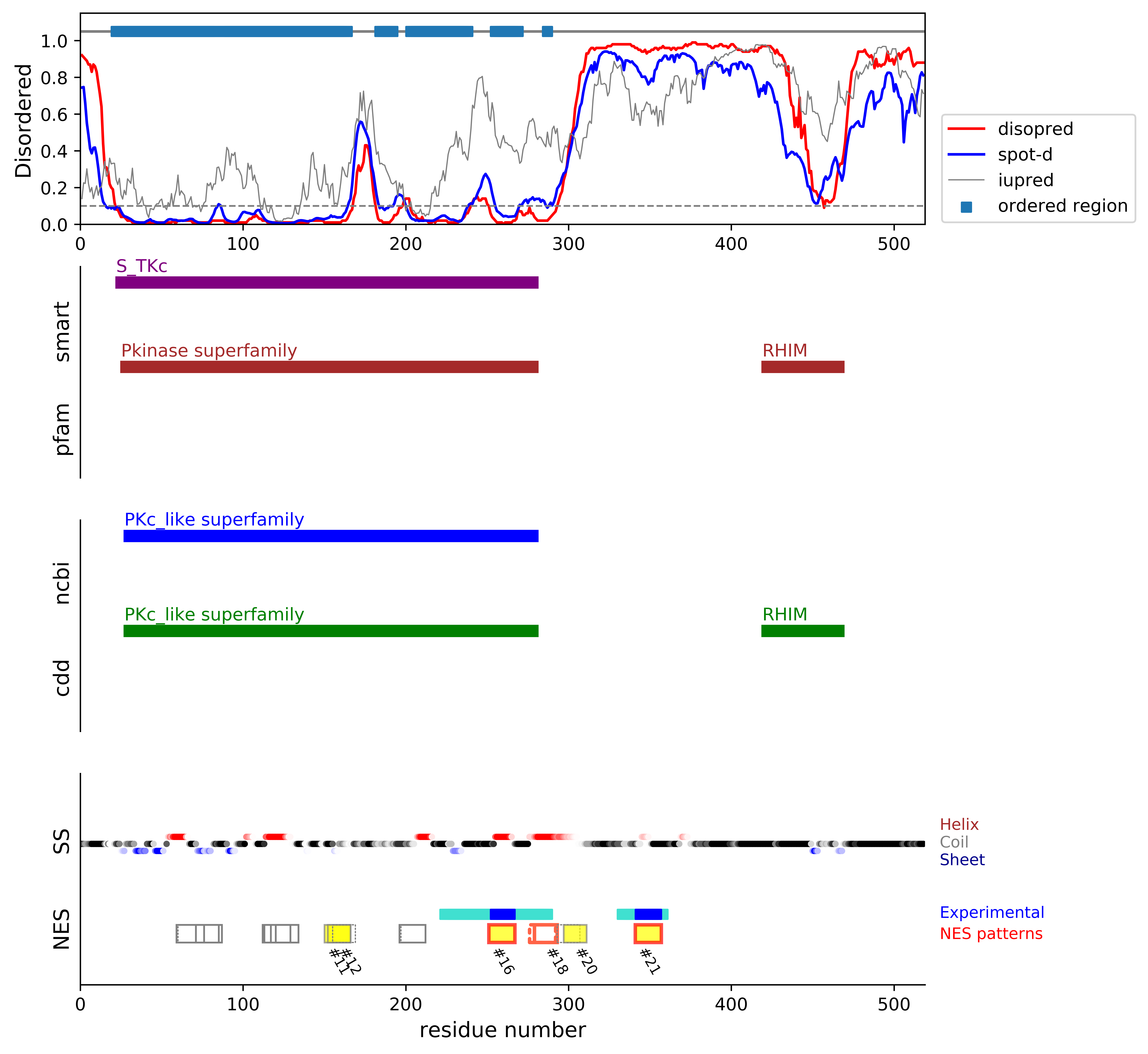

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_O | Q9Y572 | 59 | REVKAMASLDNEFVLRL | HHHHHHHHCCCCCEEEE | c1c-AT-4 | multi-selected | 0.015 | 0.019 | 0.133 | ORD | MID|PKc_like superfamily; | 0.0 |
| 2 | fp_O | Q9Y572 | 60 | EVKAMASLDNEFVLRL | HHHHHHHCCCCCEEEE | c1d-5 | multi | 0.016 | 0.019 | 0.13 | ORD | MID|PKc_like superfamily; | 0.0 |
| 3 | fp_beta_O | Q9Y572 | 71 | FVLRLEGVIEKVNWDQ | CEEEEEEEEECCCCCC | c1aR-4 | multi-selected | 0.014 | 0.037 | 0.18 | ORD | MID|PKc_like superfamily; | 0.857 |
| 4 | fp_beta_O | Q9Y572 | 71 | FVLRLEGVIEKVNW | CEEEEEEEEECCCC | c3-4 | multi-selected | 0.013 | 0.027 | 0.164 | ORD | MID|PKc_like superfamily; | 1.0 |
| 5 | fp_O | Q9Y572 | 112 | RPWPLLCRLLKEVVLGM | CCHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.012 | 0.013 | 0.052 | ORD | MID|PKc_like superfamily; | 0.0 |
| 6 | fp_O | Q9Y572 | 113 | PWPLLCRLLKEVVLGM | CHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.011 | 0.011 | 0.05 | ORD | MID|PKc_like superfamily; | 0.0 |
| 7 | fp_O | Q9Y572 | 113 | PWPLLCRLLKEVVLGM | CHHHHHHHHHHHHHHH | c1d-4 | multi | 0.011 | 0.011 | 0.05 | ORD | MID|PKc_like superfamily; | 0.0 |
| 8 | fp_O | Q9Y572 | 117 | LCRLLKEVVLGMFYLHD | HHHHHHHHHHHHHCCCC | c1c-4 | multi-selected | 0.01 | 0.008 | 0.042 | ORD | MID|PKc_like superfamily; | 0.0 |
| 9 | fp_O | Q9Y572 | 120 | LLKEVVLGMFYLHD | HHHHHHHHHHCCCC | c2-5 | multi-selected | 0.01 | 0.007 | 0.036 | ORD | MID|PKc_like superfamily; | 0.0 |
| 10 | fp_O | Q9Y572 | 120 | LLKEVVLGMFYLHD | HHHHHHHHHHCCCC | c3-4 | multi-selected | 0.01 | 0.007 | 0.036 | ORD | MID|PKc_like superfamily; | 0.0 |
| 11 | fp_D | Q9Y572 | 150 | LDPELHVKLADFGLST | CCCCCCEEECCCCCCC | c1a-4 | multi-selected | 0.016 | 0.049 | 0.22 | boundary | MID|PKc_like superfamily; | 0.429 |
| 12 | fp_D | Q9Y572 | 152 | PELHVKLADFGLST | CCCCEEECCCCCCC | c2-4 | multi-selected | 0.016 | 0.052 | 0.212 | boundary | MID|PKc_like superfamily; | 0.429 |
| 13 | fp_D | Q9Y572 | 155 | HVKLADFGLSTFQG | CEEECCCCCCCCCC | c3-AT | multi | 0.035 | 0.091 | 0.254 | boundary | MID|PKc_like superfamily; | 0.143 |
| 14 | fp_D | Q9Y572 | 196 | RKASTASDVYSFGILM | CCCCCCCCCCHHHHHH | c1a-AT-4 | multi-selected | 0.076 | 0.065 | 0.105 | boundary | MID|PKc_like superfamily; | 0.0 |
| 15 | fp_D | Q9Y572 | 197 | KASTASDVYSFGILM | CCCCCCCCCHHHHHH | c1b-AT-4 | multi | 0.075 | 0.059 | 0.097 | boundary | MID|PKc_like superfamily; | 0.0 |
| 16 | cand_D | Q9Y572 | 251 | ETPGLEGLKELMQLCW ++++*++*++*++*++ | CCCCHHHHHHHHHHHC | c1d-4 | uniq | 0.023 | 0.088 | 0.483 | boundary | boundary|PKc_like superfamily; | 0.0 |
| 17 | cand_D | Q9Y572 | 276 | FQECLPKTDEVFQMVE ++++++++++++ | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.038 | 0.124 | 0.492 | boundary | boundary|PKc_like superfamily; | 0.0 |
| 18 | cand_D | Q9Y572 | 279 | CLPKTDEVFQMVEN +++++++++ | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.033 | 0.123 | 0.476 | boundary | boundary|PKc_like superfamily; | 0.0 |
| 19 | fp_D | Q9Y572 | 293 | NMNAAVSTVKDFLS | HHHHHHHHHHHHHC | c3-AT | multi | 0.381 | 0.364 | 0.432 | DISO | boundary|PKc_like superfamily; | 0.0 |
| 20 | fp_D | Q9Y572 | 297 | AVSTVKDFLSQLRS | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.579 | 0.499 | 0.437 | DISO | boundary|PKc_like superfamily; | 0.0 |
| 21 | cand_D | Q9Y572 | 341 | NDVMVSEWLNKLNLEE +++*++++*++*+*++ | CCCCHHHHHCCCCCCC | c1a-4 | uniq | 0.949 | 0.821 | 0.616 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment