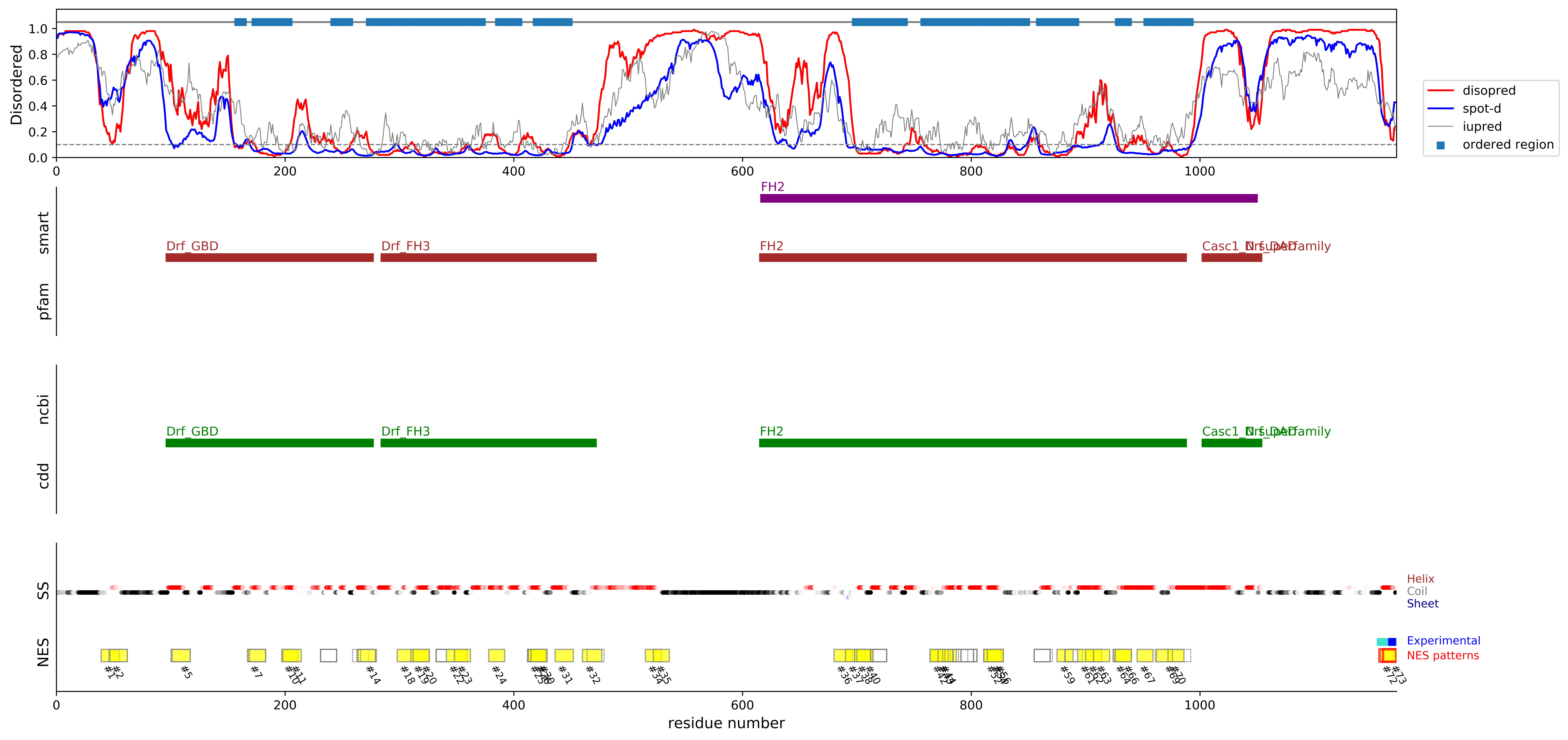

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9Z207 | 39 | LHLNIRTLTDDMLDKF | CCCCCCCCCHHHHHHH | c1aR-4 | uniq | 0.216 | 0.463 | 0.511 | DISO | 0.0 | |
| 2 | fp_D | Q9Z207 | 46 | LTDDMLDKFASIRIPG | CCHHHHHHHHCCCCCC | c1a-4 | multi-selected | 0.271 | 0.514 | 0.508 | DISO | 0.0 | |
| 3 | fp_D | Q9Z207 | 47 | TDDMLDKFASIRIPG | CHHHHHHHHCCCCCC | c1b-4 | multi | 0.276 | 0.518 | 0.509 | DISO | 0.0 | |
| 4 | fp_D | Q9Z207 | 100 | EVLKLFEKMMEDMNLNE | HHHHHHHHHHHHCCCCH | c1c-5 | multi | 0.436 | 0.118 | 0.486 | DISO | boundary|Drf_GBD; | 0.0 |
| 5 | fp_D | Q9Z207 | 101 | VLKLFEKMMEDMNLNE | HHHHHHHHHHHCCCCH | c1a-5 | multi-selected | 0.418 | 0.118 | 0.483 | DISO | boundary|Drf_GBD; | 0.0 |
| 6 | fp_D | Q9Z207 | 101 | VLKLFEKMMEDMNLNE | HHHHHHHHHHHCCCCH | c1d-5 | multi | 0.418 | 0.118 | 0.483 | DISO | boundary|Drf_GBD; | 0.0 |
| 7 | fp_D | Q9Z207 | 167 | TDERLFTYLESLRVSL | CCHHHHHHHHHHHHHC | c1a-4 | multi-selected | 0.079 | 0.071 | 0.151 | boundary | MID|Drf_GBD; | 0.0 |
| 8 | fp_D | Q9Z207 | 168 | DERLFTYLESLRVSL | CHHHHHHHHHHHHHC | c1b-4 | multi | 0.076 | 0.067 | 0.15 | boundary | MID|Drf_GBD; | 0.0 |
| 9 | fp_D | Q9Z207 | 169 | ERLFTYLESLRVSL | HHHHHHHHHHHHHC | c2-AT-4 | multi | 0.071 | 0.062 | 0.154 | boundary | MID|Drf_GBD; | 0.0 |
| 10 | fp_D | Q9Z207 | 197 | GLGLLLDILEKLIN | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.141 | 0.04 | 0.07 | boundary | MID|Drf_GBD; | 0.0 |
| 11 | fp_D | Q9Z207 | 198 | LGLLLDILEKLINGQI | HHHHHHHHHHHHCCCC | c1aR-4 | multi-selected | 0.199 | 0.063 | 0.092 | boundary | MID|Drf_GBD; | 0.0 |
| 12 | fp_D | Q9Z207 | 231 | ALMNTQYGLERIMS | HHHCCHHHHHHHHC | c3-AT | uniq | 0.128 | 0.048 | 0.148 | boundary | MID|Drf_GBD; | 0.0 |
| 13 | fp_D | Q9Z207 | 259 | RQPAMMADVVKLLS | CCCHHHHHHHHHHH | c2-AT-4 | multi | 0.166 | 0.026 | 0.122 | __ | boundary|Drf_GBD; boundary|Drf_FH3; | 0.0 |
| 14 | fp_D | Q9Z207 | 263 | MMADVVKLLSAVCIVG | HHHHHHHHHHHHHCCC | c1a-5 | multi-selected | 0.131 | 0.018 | 0.075 | boundary | boundary|Drf_GBD; boundary|Drf_FH3; | 0.0 |
| 15 | fp_D | Q9Z207 | 263 | MMADVVKLLSAVCIVG | HHHHHHHHHHHHHCCC | c1d-5 | multi | 0.131 | 0.018 | 0.075 | boundary | boundary|Drf_GBD; boundary|Drf_FH3; | 0.0 |
| 16 | fp_D | Q9Z207 | 264 | MADVVKLLSAVCIVG | HHHHHHHHHHHHCCC | c1b-4 | multi | 0.125 | 0.017 | 0.064 | __ | boundary|Drf_GBD; boundary|Drf_FH3; | 0.0 |
| 17 | fp_D | Q9Z207 | 266 | DVVKLLSAVCIVGE | HHHHHHHHHHCCCC | c3-4 | multi | 0.111 | 0.019 | 0.047 | boundary | boundary|Drf_GBD; boundary|Drf_FH3; | 0.0 |
| 18 | fp_D | Q9Z207 | 298 | KIDRFFSIVEGLRH | CCCCHHHHHHHHCC | c3-4 | uniq | 0.074 | 0.052 | 0.128 | boundary | boundary|Drf_GBD; boundary|Drf_FH3; | 0.0 |
| 19 | fp_D | Q9Z207 | 310 | RHNSVNLQVACMQLIN | CCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.05 | 0.028 | 0.089 | boundary | boundary|Drf_GBD; MID|Drf_FH3; | 0.0 |
| 20 | fp_D | Q9Z207 | 312 | NSVNLQVACMQLIN | CCHHHHHHHHHHHH | c2-4 | multi-selected | 0.041 | 0.026 | 0.093 | __ | MID|Drf_FH3; | 0.0 |
| 21 | fp_O | Q9Z207 | 332 | DDLDFRLHLRNEFMRCG | CCHHHHHHHHHHHHHCC | c1cR-4 | uniq | 0.035 | 0.031 | 0.091 | ORD | MID|Drf_FH3; | 0.0 |
| 22 | fp_D | Q9Z207 | 341 | RNEFMRCGLKEILPNLKG | HHHHHHCCHHHHHHHHCC | c4-4 | uniq | 0.046 | 0.032 | 0.093 | boundary | MID|Drf_FH3; | 0.0 |
| 23 | fp_D | Q9Z207 | 348 | GLKEILPNLKGIKN | CHHHHHHHHCCCCC | c3-4 | uniq | 0.06 | 0.048 | 0.109 | boundary | MID|Drf_FH3; | 0.0 |
| 24 | fp_D | Q9Z207 | 378 | DLSEFFHRLEDIRA | HHHHHHHHHHHHHH | c3-4 | uniq | 0.132 | 0.032 | 0.149 | boundary | MID|Drf_FH3; | 0.0 |
| 25 | fp_D | Q9Z207 | 412 | AEGHFLSILQHLLLIRN | CHHHHHHHHHHHHHCCC | c1c-4 | multi-selected | 0.101 | 0.025 | 0.065 | boundary | MID|Drf_FH3; | 0.0 |
| 26 | fp_D | Q9Z207 | 412 | AEGHFLSILQHLLLIR | CHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.104 | 0.023 | 0.067 | boundary | MID|Drf_FH3; | 0.0 |
| 27 | fp_D | Q9Z207 | 412 | AEGHFLSILQHLLLIR | CHHHHHHHHHHHHHCC | c1d-4 | multi | 0.104 | 0.023 | 0.067 | boundary | MID|Drf_FH3; | 0.0 |
| 28 | fp_D | Q9Z207 | 413 | EGHFLSILQHLLLIR | HHHHHHHHHHHHHCC | c1b-4 | multi | 0.098 | 0.022 | 0.063 | boundary | MID|Drf_FH3; | 0.0 |
| 29 | fp_D | Q9Z207 | 413 | EGHFLSILQHLLLIRN | HHHHHHHHHHHHHCCC | c1d-4 | multi | 0.096 | 0.024 | 0.062 | boundary | MID|Drf_FH3; | 0.0 |
| 30 | fp_D | Q9Z207 | 415 | HFLSILQHLLLIRN | HHHHHHHHHHHCCC | c3-4 | multi-selected | 0.086 | 0.023 | 0.057 | boundary | MID|Drf_FH3; | 0.0 |
| 31 | fp_D | Q9Z207 | 436 | YFKLIDECVSQIVLHR | HHHHHHHHHHHHHHCC | c1a-5 | uniq | 0.053 | 0.04 | 0.119 | boundary | boundary|Drf_FH3; | 0.0 |
| 32 | fp_D | Q9Z207 | 460 | YRKRLDLDLSQFVDVCI | CCCCCCCCHHHHHHHHH | c1c-4 | multi-selected | 0.198 | 0.116 | 0.185 | boundary | boundary|Drf_FH3; | 0.0 |
| 33 | fp_D | Q9Z207 | 464 | LDLDLSQFVDVCIDQ | CCCCHHHHHHHHHHH | c1b-4 | multi | 0.25 | 0.105 | 0.155 | DISO | boundary|Drf_FH3; | 0.0 |
| 34 | fp_D | Q9Z207 | 515 | KINELQAELQAFKS | HHHHHHHHHHHHHC | c3-4 | uniq | 0.911 | 0.499 | 0.477 | DISO | 0.0 | |
| 35 | fp_D | Q9Z207 | 522 | ELQAFKSQFGALPP | HHHHHHCCCCCCCC | c3-4 | uniq | 0.946 | 0.573 | 0.476 | DISO | 0.0 | |
| 36 | fp_D | Q9Z207 | 680 | EKKVIKKRMKELKFLD | HHHHHCCCCCCEEEEC | c1a-4 | uniq | 0.781 | 0.327 | 0.295 | boundary | MID|FH2; | 0.0 |
| 37 | fp_D | Q9Z207 | 690 | ELKFLDPKIAQNLSIFL | CEEEECCCCHHHHHHHH | c1c-5 | uniq | 0.255 | 0.08 | 0.126 | boundary | MID|FH2; | 0.125 |
| 38 | fp_D | Q9Z207 | 698 | IAQNLSIFLSSFRVPY | CHHHHHHHHHCCCCCH | c1a-4 | multi-selected | 0.041 | 0.068 | 0.072 | boundary | MID|FH2; | 0.0 |
| 39 | fp_D | Q9Z207 | 698 | IAQNLSIFLSSFRVPY | CHHHHHHHHHCCCCCH | c1d-4 | multi | 0.041 | 0.068 | 0.072 | boundary | MID|FH2; | 0.0 |
| 40 | fp_D | Q9Z207 | 700 | QNLSIFLSSFRVPY | HHHHHHHHCCCCCH | c2-4 | multi-selected | 0.033 | 0.066 | 0.07 | boundary | MID|FH2; | 0.0 |
| 41 | fp_D | Q9Z207 | 712 | PYEKIRTMILEVDE | CHHHHHHHHHHHHH | c2-AT-4 | uniq | 0.039 | 0.063 | 0.176 | boundary | MID|FH2; | 0.0 |
| 42 | fp_D | Q9Z207 | 764 | QFAVVMSNVKRLRP | CHHHHCCCCCCHHH | c3-4 | multi-selected | 0.045 | 0.027 | 0.15 | boundary | MID|FH2; | 0.0 |
| 43 | fp_D | Q9Z207 | 764 | QFAVVMSNVKRLRPRLSA | CHHHHCCCCCCHHHHHHH | c4-5 | multi-selected | 0.039 | 0.025 | 0.135 | boundary | MID|FH2; | 0.0 |
| 44 | fp_D | Q9Z207 | 771 | NVKRLRPRLSAILFKL | CCCCHHHHHHHHHHHH | c1a-5 | multi-selected | 0.026 | 0.024 | 0.097 | __ | MID|FH2; | 0.0 |
| 45 | fp_O | Q9Z207 | 775 | LRPRLSAILFKLQFEE | HHHHHHHHHHHHHHHH | c1a-4 | multi | 0.019 | 0.022 | 0.082 | ORD | MID|FH2; | 0.0 |
| 46 | fp_O | Q9Z207 | 775 | LRPRLSAILFKLQFEE | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.019 | 0.022 | 0.082 | ORD | MID|FH2; | 0.0 |
| 47 | fp_O | Q9Z207 | 775 | LRPRLSAILFKLQF | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.019 | 0.022 | 0.083 | ORD | MID|FH2; | 0.0 |
| 48 | fp_O | Q9Z207 | 777 | PRLSAILFKLQFEE | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.016 | 0.021 | 0.087 | ORD | MID|FH2; | 0.0 |
| 49 | fp_O | Q9Z207 | 780 | SAILFKLQFEEQVNNIK | HHHHHHHHHHHHHHHHH | c1cR-5 | multi | 0.017 | 0.024 | 0.108 | ORD | MID|FH2; | 0.0 |
| 50 | fp_O | Q9Z207 | 784 | FKLQFEEQVNNIKPDIMA | HHHHHHHHHHHHHHHHHH | c4-4 | multi-selected | 0.025 | 0.026 | 0.156 | ORD | MID|FH2; | 0.0 |
| 51 | fp_O | Q9Z207 | 791 | QVNNIKPDIMAVST | HHHHHHHHHHHHHH | c3-4 | uniq | 0.041 | 0.028 | 0.215 | ORD | MID|FH2; | 0.0 |
| 52 | fp_D | Q9Z207 | 811 | KSKGFSKLLELVLLMG | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.034 | 0.023 | 0.101 | boundary | MID|FH2; | 0.0 |
| 53 | fp_D | Q9Z207 | 811 | KSKGFSKLLELVLLMG | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.034 | 0.023 | 0.101 | boundary | MID|FH2; | 0.0 |
| 54 | fp_D | Q9Z207 | 811 | KSKGFSKLLELVLLMGN | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.034 | 0.022 | 0.106 | boundary | MID|FH2; | 0.0 |
| 55 | fp_D | Q9Z207 | 811 | KSKGFSKLLELVLLM | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.034 | 0.023 | 0.097 | boundary | MID|FH2; | 0.0 |
| 56 | fp_D | Q9Z207 | 814 | GFSKLLELVLLMGN | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.025 | 0.019 | 0.102 | boundary | MID|FH2; | 0.0 |
| 57 | fp_D | Q9Z207 | 855 | ADQKTTLLHFLVDVCE | CCCCCCHHHHHHHHHH | c1d-AT-4 | multi | 0.085 | 0.071 | 0.14 | boundary | MID|FH2; | 0.0 |
| 58 | fp_D | Q9Z207 | 855 | ADQKTTLLHFLVDV | CCCCCCHHHHHHHH | c2-AT-4 | multi-selected | 0.094 | 0.078 | 0.156 | boundary | MID|FH2; | 0.0 |
| 59 | fp_D | Q9Z207 | 875 | DILHFVDDLAHLDK | CCCCCHHHCCCHHH | c3-4 | uniq | 0.026 | 0.049 | 0.119 | boundary | MID|FH2; | 0.0 |
| 60 | fp_D | Q9Z207 | 882 | DLAHLDKASRVSVEM | HCCCHHHHHCCCHHH | c1b-AT-5 | uniq | 0.097 | 0.075 | 0.192 | boundary | MID|FH2; | 0.0 |
| 61 | fp_D | Q9Z207 | 893 | SVEMLEKNVKQMGR | CHHHHHHHHHHHHH | c3-4 | uniq | 0.242 | 0.082 | 0.347 | boundary | MID|FH2; | 0.0 |
| 62 | fp_D | Q9Z207 | 900 | NVKQMGRQLQQLEK | HHHHHHHHHHHHHH | c3-4 | uniq | 0.369 | 0.089 | 0.419 | boundary | MID|FH2; | 0.0 |
| 63 | fp_D | Q9Z207 | 907 | QLQQLEKNLETFPP | HHHHHHHHHHCCCC | c3-4 | uniq | 0.454 | 0.132 | 0.468 | boundary | MID|FH2; | 0.0 |
| 64 | fp_D | Q9Z207 | 924 | LHDKFVIKMSSFVISA | CCCHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.128 | 0.067 | 0.193 | boundary | MID|FH2; | 0.0 |
| 65 | fp_D | Q9Z207 | 925 | HDKFVIKMSSFVISA | CCHHHHHHHHHHHHH | c1b-4 | multi | 0.114 | 0.059 | 0.181 | boundary | MID|FH2; | 0.0 |
| 66 | fp_D | Q9Z207 | 926 | DKFVIKMSSFVISA | CHHHHHHHHHHHHH | c2-4 | multi-selected | 0.102 | 0.052 | 0.177 | boundary | MID|FH2; | 0.0 |
| 67 | fp_D | Q9Z207 | 945 | KLSTLLGSMTQLYQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.112 | 0.028 | 0.199 | boundary | MID|FH2; | 0.0 |
| 68 | fp_D | Q9Z207 | 961 | MGYYAVDMKKVSVEE | HHHHCCCCCCCCHHH | c1b-AT-4 | multi | 0.068 | 0.045 | 0.139 | boundary | boundary|FH2; | 0.0 |
| 69 | fp_D | Q9Z207 | 962 | GYYAVDMKKVSVEE | HHHCCCCCCCCHHH | c2-4 | multi-selected | 0.071 | 0.046 | 0.137 | boundary | boundary|FH2; | 0.0 |
| 70 | fp_D | Q9Z207 | 972 | SVEEFFNDLNNFRT | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.051 | 0.055 | 0.183 | boundary | boundary|FH2; | 0.0 |
| 71 | fp_D | Q9Z207 | 976 | FFNDLNNFRTSFMLAL | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.033 | 0.062 | 0.224 | boundary | boundary|FH2; | 0.0 |
| 72 | cand_D | Q9Z207 | 1157 | SVPEVEALLARLRA ++++++++++*++ | CHHHHHHHHHHHHC | c3-4 | multi-selected | 0.372 | 0.321 | 0.387 | DISO | 0.0 | |
| 73 | cand_D | Q9Z207 | 1160 | EVEALLARLRALXX ++++++++*+++ | HHHHHHHHHHCC | c3-4-Ct | multi-selected | 0.258 | 0.305 | 0.366 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment