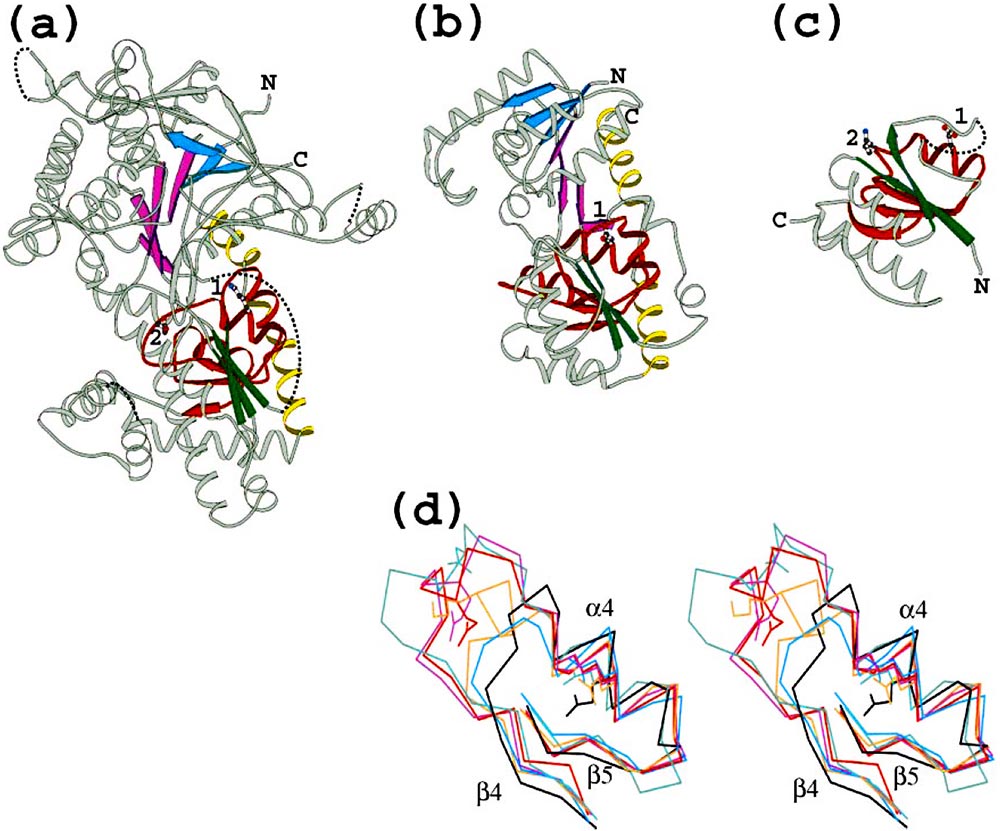
Structures of diverse O-linked GlcNAc transferase homologs: (a) yeast glycogen phosphorylase (PDB 1ygpA42), (b) bacterial glycosyl transferase MurG (PDB 1f0kA24), (c) Thermus thermophilus ribosomal protein S2 (PDB 1fjfB39). β-Strands in the common structural core are color coded . Additional α-helices near the conserved GPGTF motif in domain II are colored red. Two conserved helices at the extreme C-terminus of 1ygpA and 1f0kA are colored yellow. Side chains of conserved residues 1 and 2 are indicated by numbers. Coordinate gaps in the PDB file and some deleted non-homologous regions are connected by dotted lines to facilitate chain tracing. Figures were composed with Bobscript. (d) Stereo view of superimposition of the GPGTF (glycogen phosphorylase/glycosyl transferase) motif region (IIβ4 to IIβ5) for six PDB structures. Portions of PDB structures of T4 phage β-glucosyltransferase (green, 1c3jA34), bacterial maltodextrin phosphorylase (red, 1e4oA22), bacterial MurG (black, 1f0kA24), bacterial UDP-GlcNAc 2-epimerase (blue, 1f6dA23), T. thermophilus ribosomal protein S2 (brown, 1fjfB39), yeast glycogen phosphorylase (purple, 1ygpA42). The orientation of the structural fragment is identical to the orientation of the complete structures in (a)-(c). Conserved side chains 1 and 2 are explicitly shown.