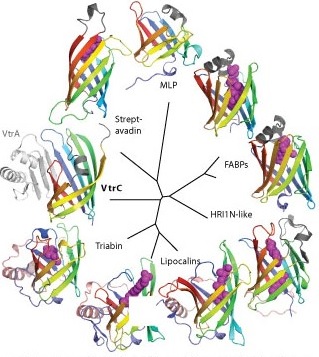
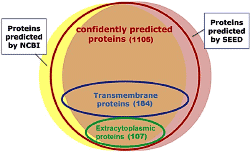
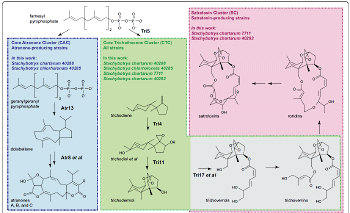
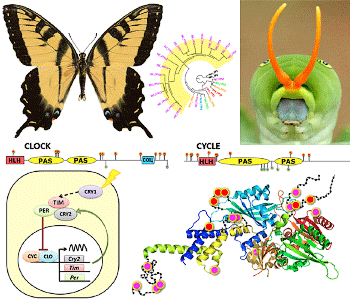
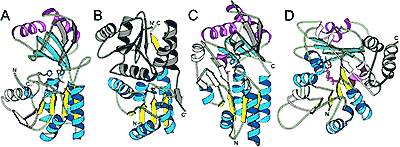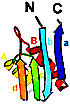Research
"Nothing in biology makes sense
except in the light of evolution."
T. Dobzhansky (1900-1975)
We develop and use theoretical methods to study proteins, genomes and organisms
We work at the interface of biology, computer science, mathematics and physics. Our group specializes in computational biology of proteins and genomes and combines sequence and structure analysis with evolutionary considerations to facilitate discoveries of biological significance. Two major directions are pursued:
|
This duality, i.e., a combination of methods development with biological applications is beneficial to both directions, as we frequently find that existing approaches do not give satisfactory answers to specific biological questions. Thus we develop new methods to fill the void. The availability of experts biologists in our group to validate the results of new approaches, in turn, stimulates methods development.
• Introduction •
What is the most important unsolved problem in computational biology of proteins? Apparently, it is protein energetics. 1) The protein folding problem (i.e. prediction of spatial structure from sequence; 2) the precise modeling of interactions between proteins and of proteins with other molecules; and 3) the quantitative understanding of enzyme catalysis — are all various incarnations of the same challenge. Despite significant achievements in the field, an exact solution from the physics perspective is still far from reach.
Bioinformatics approaches offer practical shortcuts to these problems. Deduction of protein properties (i.e. 3D structure or function) by homology to proteins with known properties has been the most successful application. For this method to reach its full potential, the following steps should be perfected. 1) find homologous proteins, i.e. do a database search; 2) compare them to the protein of interest, e.g. make an alignment; 3) decide on the boundaries of property transfer by similarity, i.e. at what level of similarity a property is shared between homologs, and thus can be deduced without experimental characterization. Most projects in the lab deal with these questions.
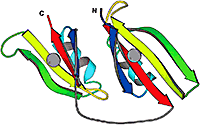
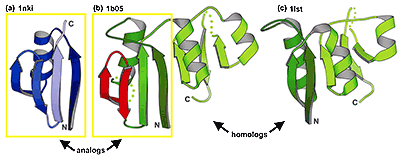
Homology and evolution are the central themes of our research. By homology we mean similarity caused by common ancestry, not just any similarity. Proteins are homologous if they originated from a common ancestor. Similarity caused by other reasons, e.g. structural constraints on 3D packing, is termed analogy. When similarity is weak, distinguishing between the two scenarios, homology vs. analogy, is challenging. We are working on this problem.
• Main Research Directions •
The long-term objective of our research is to classify available protein sequence-structure data into a biologically relevant, hierarchical system analogous to the one currently used in zoology and botany, and to provide computational tools to establish and maintain this classification. Since sequence and structural similarities usually imply functional similarity, such classification would provide an indispensable tool for biologists to aid in experimental design. Applying a combination of various approaches is usually best for addressing complex problems. Thus the questions we pursue are quite diverse and can be summarized as follows.
Our publications give more specific ideas about research directions. Additionally, we are involved in many collaborations to study individual protein families, predict properties of proteins, help interpret the effects of clinical mutations and assist in data analysis–driven experimental design. Below are short descriptions of projects and main results.
Evolutionary classification of proteins up
Protein structure classification is necessary to comprehend the rapidly growing structural data for better understanding of protein evolution and sequence-structure-function relationships. Moving towards the goal of classifying all proteins, we focused on small domains: zinc-fingers and disulfide-rich modules, and several other protein classes (e.g. kinases and thioredoxin fold proteins).
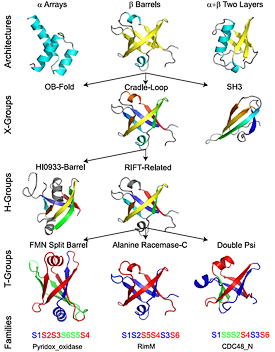
ECOD is a hierarchical evolutionary classification of all proteins with experimentally determined spatial structures that we developed and implemented as an interactive and updatable on-line database. ECOD (Evolutionary Classification of Domains) is distinct from other structural classifications in that it groups domains primarily by evolutionary relationships (homology), rather than topology (or "fold"). This distinction highlights cases of homology between domains of differing topology to aid in understanding of protein structure evolution. ECOD uniquely emphasizes distantly related homologs that are difficult to detect and thus catalogs the largest number of evolutionary links among structural domain classifications. Placing distant homologs together underscores the ancestral similarities of these proteins and draws attention to the most important regions of sequence and structure, as well as conserved functional sites. ECOD also recognizes closer sequence-based relationships between protein domains. Over 100,000 protein structures are classified in ECOD into 11,000 sequence families clustered into nearly 3,000 evolutionary groups. The classification is assisted by an automated pipeline that quickly and consistently classifies weekly releases of PDB structures and allows for continual updates. This synchronization with PDB uniquely distinguishes ECOD among all protein classifications. The results are available as an online database at http://prodata.swmed.edu/ecod PDF
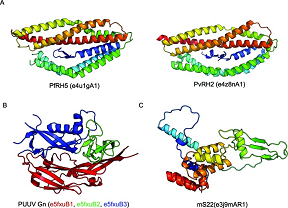
As of 2017, we have completed over 123 weekly updates and release of ECOD. We have added representative domain sets to our suite of distributable files, as well as generated PDB-style structure files for these representative domain sets. Furthermore, these weekly updates have allowed ECOD curators to identify and curate multiple previously unidentified homologous links between PDB structures. Reticulocyte-binding protein homolog PfRH5 (4U1G) was used as seed for a novel homologous group, wherein subsequent viral homologs were classified. The increase in deposited cryoEM structures has led to a increase in novel domains identified in these structures. A mitoribosomal subunit ms22 was split into 3 domains by manual curators, 2 of which had no observable homology to previously observed domains. This sustained 18-month long period of well-curated updates illustrates the long-term sustainability of the ECOD mixed manual/automatic curation approach. PDF
Protein families are groups of highly sequence-similar proteins sharing similar functions. Multiple sequence databases of these proteins exist. We generated a set of new protein families to classify those proteins whose structure had been determined, but whose domains could not be assigned to an existing protein family. Moreover, a protein structure can reveal details about domain structure that are not obvious from a sequence-based point of view. Domain definitions can markedly differ between a database derived from protein structure and a database based on families of protein s equences. Where possible, we have used the novel protein domain architectures cataloged in ECOD to derive more finely detailed protein family definitions. Using a novel workflow for generation of families, we found that about 30% of ECOD families are equivalent to known sequence families (i.e from Pfam), an additional 30% are overlap significantly with a Pfam family but the domain boundaries involved are different, and the remainder being some variety of previously uncataloged protein family. PDF
The classical Rossmann fold, also known as a doubly-wound three layer/sandwich, consists of two-units (321456 topology) that form a single parallel sheet flanked by alpha-helices on both sides and contain a characteristic crossover between strands 3 and 4. We defined its core minimal Rossmann-like motif (RLM) unit of three beta-strands flanked by two alpha-helices and found all known protein structures containing the RLM. We show that RLM enzymes function predominantly in metabolism, covering 38% of reference metabolic pathways. We find that closely related RLM enzyme families can catalyze different reaction chemistries using similar folds. Alternatively, different RLM folds can converge on catalyzing the same reactions. We showed that RLM enzymes utilize ligands from 20 chemical superclasses of organic and inorganic compounds. Homologous RLM domains can exhibit diverging active sites that accommodate alternate ligands, but with similar binding modes. The Rossmann fold is considered one of the most ancient folds, utilizing iron-sulfur clusters as cofactors and being the part of ancient energy metabolism, the Wood-Ljungdahl pathway, used by LUCA. Our data suggests that the top three disease categories with mutations in RLM proteins are diseases of endocrine system, nervous system and developmental anomalies. PDF1 PDF2
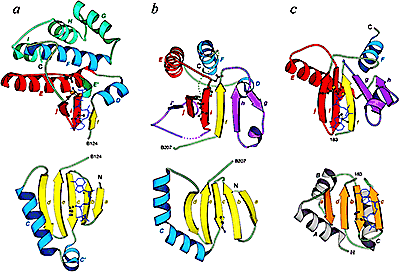
Disulfide-rich domains are small protein domains whose global folds are stabilized primarily by the formation of disulfide bonds and, to a much lesser extent, by secondary structure and hydrophobic interactions. Disulfide-rich domains perform a wide variety of roles functioning as growth factors, toxins, enzyme inhibitors, hormones, pheromones, allergens, etc. These domains are commonly found both as independent (single-domain) proteins and as domains within larger polypeptides. We compiled a comprehensive structural classification of approximately 3000 small, disulfide-rich protein domains. We find that these domains can be arranged into 41 fold groups on the basis of structural similarity. Our fold groups, which describe broader structural relationships than existing groupings of these domains, bring together representatives with previously unacknowledged similarities; 18 of the 41 fold groups include domains from several SCOP folds. Within the fold groups, the domains are assembled into families of homologs. We define 98 families of disulfide-rich domains, some of which include newly detected homologs, particularly among knottin-like domains. On the basis of this classification, we have examined cases of convergent and divergent evolution of functions performed by disulfide-rich proteins. Disulfide bonding patterns in these domains are also evaluated. Reducible disulfide bonding patterns are much less frequent, while symmetric disulfide bonding patterns are more common than expected from random considerations. PDF
Zinc fingers are small protein domains in which zinc plays a structural role contributing to the stability of the domain. Zinc fingers are structurally diverse and are present among proteins that perform a broad range of functions in various cellular processes, such as replication and repair, transcription and translation, metabolism and signaling, cell proliferation and apoptosis. Zinc fingers typically function as interaction modules and bind to a wide variety of compounds, such as nucleic acids, proteins and small molecules. Here we present a comprehensive classification of zinc finger spatial structures. We find that each available zinc finger structure can be placed into one of eight fold groups that we define based on the structural properties in the vicinity of the zinc-binding site. Three of these fold groups comprise the majority of zinc fingers, namely, C2H2-like finger, treble clef finger and the zinc ribbon. Evolutionary relatedness of proteins within fold groups is not implied, but each group is divided into families of potential homologs. We compare our classification to existing groupings of zinc fingers and find that we define more encompassing fold groups, which bring together proteins whose similarities have previously remained unappreciated. We analyze functional properties of different zinc fingers and overlay them onto our classification. The classification helps in understanding the relationship between the structure, function and evolutionary history of these domains. The results are available as an online database of zinc finger structures. PDF
Kinases are ubiquitous enzymes that catalyze the phosphoryl transfer reaction from a phosphate donor (usually ATP) to a receptor substrate. Although all kinases catalyze essentially the same phosphoryl transfer reaction, they display remarkable diversity in their substrate specificity, structure, and the pathways in which they participate. In order to learn the relationship between structural fold and functional specificities in kinases, we have done a comprehensive survey of all available kinase sequences (>17,000) and classified them into 30 distinct families based on sequence similarities. Of these families, 19, covering nearly 98% of all sequences, fall into seven general structural folds for which three-dimensional structures are known. These fold groups include some of the most widespread protein folds, such as Rossmann fold, ferredoxin fold, ribonuclease H fold, and TIM beta/alpha-barrel. On the basis of this classification system, we examined the shared substrate binding and catalytic mechanisms as well as variations of these mechanisms in the same fold groups. Cases of convergent evolution of identical kinase activities occurring in different folds were identified. Three years later, a comprehensive update of the classification of all available kinases was carried out. This survey presents a complete global picture of this large functional class of proteins and confirms the soundness of our initial kinase classification scheme. The new survey found the total number of kinase sequences in the protein database has increased more than three-fold (from 17,310 to 59,402), and the number of determined kinase structures increased two-fold (from 359 to 702) in the past three years. However, the framework of the original two-tier classification scheme (in families and fold groups) remains sufficient to describe all available kinases. Overall, the kinase sequences were classified into 25 families of homologous proteins, wherein 22 families (approximately 98.8% of all sequences) for which three-dimensional structures are known fall into 10 fold groups. These fold groups not only include some of the most widely spread proteins folds, such as the Rossmann-like fold, ferredoxin-like fold, TIM-barrel fold, and antiparallel beta-barrel fold, but also all major classes (all alpha, all beta, alpha+beta, alpha/beta) of protein structures. Fold predictions are made for remaining kinase families without a close homolog with solved structure. We also highlight two novel kinase structural folds, riboflavin kinase and dihydroxyacetone kinase, which have recently been characterized. Two protein families previously annotated as kinases are removed from the classification based on new experimental data. CONCLUSION: Structural annotations of all kinase families are now revealed, including fold descriptions for all globular kinases, making this the first large functional class of proteins with a comprehensive structural annotation. Potential uses for this classification include deduction of protein function, structural fold, or enzymatic mechanism of poorly studied or newly discovered kinases based on proteins in the same family. PDF1 PDF2
Thioredoxins are important proteins that ubiquitously regulate cellular redox status and various other crucial functions. We define the thioredoxin-like fold using the structure consensus of thioredoxin homologs and consider all circular permutations of the fold. The search for thioredoxin-like fold proteins in the PDB database identified 723 protein domains. These domains are grouped into eleven evolutionary families based on combined sequence, structural, and functional evidence. Analysis of the protein-ligand structure complexes reveals two major active site locations for the thioredoxin-like proteins. Comparison to existing structure classifications reveals that our thioredoxin-like fold group is broader and more inclusive, unifying proteins from five SCOP folds, five CATH topologies and seven DALI domain dictionary globular folding topologies. Considering these structurally similar domains together sheds new light on the relationships between sequence, structure, function and evolution of thioredoxins. PDF
FlyXCDB is a resource for Drosophila cell surface and secreted proteins and their extracellular domains. Genomes of metazoan organisms possess a large number of genes encoding cell surface and secreted (CSS) proteins that carry out crucial functions in cell adhesion and communication, signal transduction, extracellular matrix establishment, nutrient digestion and uptake, immunity, and developmental processes. We developed the FlyXCDB database that provides a comprehensive resource to investigate extracellular (XC) domains in CSS proteins of Drosophila melanogaster, the most studied insect model organism in various aspects of animal biology. More than 300 Drosophila XC domains were discovered in Drosophila CSS proteins encoded by over 2500 genes through analyses of computational predictions of signal peptide, transmembrane (TM) segment, and GPI-anchor signal sequence, profile-based sequence similarity searches, gene ontology, and literature. These domains were classified into six classes mainly based on their molecular functions, including protein-protein interactions (class P), signaling molecules (class S), binding of non-protein molecules or groups (class B), enzyme homologs (class E), enzyme regulation and inhibition (class R), and unknown molecular function (class U). Main cellular functions such as cell adhesion, cell signaling, and extracellular matrix composition were described for the most abundant domains in each functional class. We assigned cell membrane topology categories (E, secreted; S, type I/III single-pass TM; T, type II single-pass TM; M, multi-pass TM; and G, GPI-anchored) to the products of genes with XC domains and investigated their regulation by mechanisms such as alternative splicing and stop codon readthrough. PDF
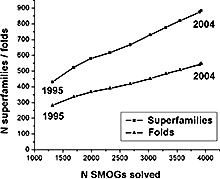
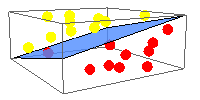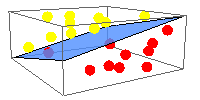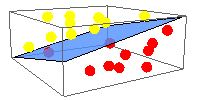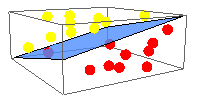
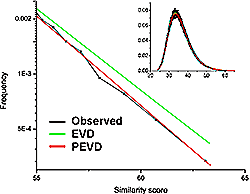
Growth of superfamilies and folds with solved 3D structures: Growth rate remains approximately linear despite the exponential growth in the number of solved structures.
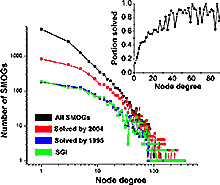
Highly connected sequence families are more likely to be solved. Inset: fraction of families with solved structure as a function of number of sequence similarity links.
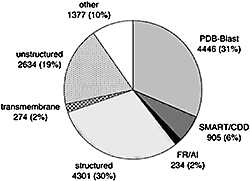
As tertiary structure is currently available only for a fraction of known protein families, it is important to assess what parts of sequence space have been structurally characterized. We consider protein domains whose structure can be predicted by sequence similarity to proteins with solved structure and address the following questions. Do these domains represent an unbiased random sample of all sequence families? Do targets solved by structural genomic initiatives (SGI) provide such a sample? What are approximate total numbers of structure-based superfamilies and folds among soluble globular domains? To make these assessments, we combine two approaches: (i) sequence analysis and homology-based structure prediction for proteins from complete genomes; and (ii) monitoring dynamics of the assigned structure set in time, with the accumulation of experimentally solved structures. In the Clusters of Orthologous Groups (COG) database, we map the growing population of structurally characterized domain families onto the network of sequence-based connections between domains. This mapping reveals a systematic bias suggesting that target families for structure determination tend to be located in highly populated areas of sequence space. In contrast, the subset of domains whose structure is initially inferred by SGI is similar to a random sample from the whole population. To accommodate for the observed bias, we propose a new non-parametric approach to the estimation of the total numbers of structural superfamilies and folds, which does not rely on a specific model of the sampling process. Based on dynamics of robust distribution-based parameters in the growing set of structure predictions, we estimate the total numbers of superfamilies and folds among soluble globular proteins in the COG database. The set of currently solved protein structures allows for structure prediction in approximately a third of sequence-based domain families. The choice of targets for structure determination is biased towards domains with many sequence-based homologs. The growing SGI output in the future should further contribute to the reduction of this bias. The total number of structural superfamilies and folds in the COG database are estimated as approximately 4000 and approximately 1700. These numbers are respectively four and three times higher than the numbers of superfamilies and folds that can currently be assigned to COG proteins. PDF
Homology vs. analogy, divergence vs. convergence up
A natural way to study protein sequence, structure, and function is to put them in the context of evolution. Homologs inherit similarities from their common ancestor, while analogs converge to similar structures due to a limited number of energetically favorable ways to pack secondary structural elements. Using novel strategies, we assembled two reliable databases of homologs and analogs.
MALIDUP (manual alignments of duplicated domains), a database of 241 pairwise structure alignments for homologous domains originated by internal duplication within the same polypeptide chain. Since duplicated domains within a protein frequently diverge in function and thus in sequence, this would be the first database of structurally similar homologs that is not strongly biased by sequence or functional similarity. Our manual alignments in most cases agree with the automatic structural alignments generated by several commonly used programs. This carefully constructed database could be used in studies on protein evolution and as a reference for testing structure alignment programs. PDF
MALISAM (manual alignments for structurally analogous motifs) represents the first database containing pairs of structural analogs and their alignments. To find reliable analogs, we developed an approach based on three ideas. First, an insertion together with a part of the evolutionary core of one domain family (a hybrid motif) is analogous to a similar motif contained within the core of another domain family. Second, a motif at an interface, formed by secondary structural elements (SSEs) contributed by two or more domains or subunits contacting along that interface, is analogous to a similar motif present in the core of a single domain. Third, an artificial protein obtained through selection from random peptides or in sequence design experiments not biased by sequences of a particular homologous family, is analogous to a structurally similar natural protein. Each analogous pair is superimposed and aligned manually, as well as by several commonly used programs. Applications of this database may range from protein evolution studies, e.g. development of remote homology inference tools and discriminators between homologs and analogs, to protein-folding research, since in the absence of evolutionary reasons, similarity between proteins is caused by structural and folding constraints. PDF1 PDF2
We compare these two data sets and develop a support vector machine (SVM)-based classifier to discriminate between homologs and analogs. The classifier uses a number of well-known similarity scores. We observe that although both structure scores and sequence scores contribute to SVM performance, profile sequence scores computed based on structural alignments are the best discriminators between remote homologs and structural analogs. We apply our classifier to a representative set from the expert-constructed database, Structural Classification of Proteins (SCOP). The SVM classifier recovers 76% of the remote homologs defined as domains in the same SCOP superfamily but from different families. More importantly, we also detect interesting homologous relationships between SCOP domains from different superfamilies, folds, and even classes. PDF
Structural fold change in evolution of proteins up
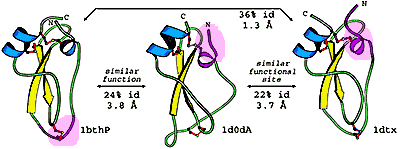
From the early days of protein structural biology, researches have been surprised by the resistance of protein spatial structures to evolutionary changes. This remarkable structural robustness combined with the limited number of available 3D structures has lead to a view that the abstract protein structure space is discrete, can be divided into a number of folds, and protein evolution mostly proceeds within the framework of the same fold. Today, with the rapidly increasing number of protein structures, arguably, the majority of protein structural patterns have been experimentally determined and a new view of structural continuity of folding patterns is starting to emerge. Many examples of proteins with statistically significant sequence similarity, but substantial structural differences, have been documented. Such phenomenon demonstrates the evolutionary bridges between structurally different proteins and profoundly influences our understanding of protein structure evolution. On one hand, the notion that protein structures are evolutionarily plastic and changeable has important applications in protein design and opens new frontiers in engineering proteins that possess desired functional properties, such as a possibility to create proteins with condition-dependent folds. On the other hand, the existence of proteins with similar sequences but different structures hinders homology modeling methods; therefore our ability to detect such cases from sequence is crucial. To study the mechanisms and paths of protein fold change in evolution, we undertook comprehensive comparative analysis of protein sequences and structures, and catalogued the instances of potentially homologous proteins with significant structural differences. Our work revealed that, although such instances are not very common, they are universally observed among proteins of all structural classes, and involve substantial structural changes and rearrangements that may be explained by both small sequence changes, such as point mutations, and large sequence rearrangements, such as non-homologous recombination. Several mechanisms such as insertions/deletions/substitutions, circular permutations, and rearrangements in b-sheet topologies account for the majority of detected structural irregularities. Fold change events are frequently correlated with the changes in oligomeric states of proteins, i.e. one of the variants is usually an oligomer, mostly frequently a dimer. It is likely that significant structual changes require additional stabilization by oligomerization. We observe that many changes, especially deteriorations, occur in auxiliary domains, not in the main functional domains. PDF1 PDF2 PDF3
To study the mechanisms and paths of protein fold change in evolution, we undertook a comprehensive comparative analysis of SCOP (Structural Classification of Proteins) domains and found domain pairs with significant sequence similarity (measured by HHsearch probability), but pronounced structural differences (measured by Dali Z-score). For all representative domain pairs, the reasons for the discordance between the sequence similarity and structural dissimilarity were studied and classified into three categories: (1) problems with the sequence or sequence alignment; (2) problems with structure or structure alignment; (3) events of interest for protein evolution and biology. We find that, on the one hand, dataset of structurally different proteins with strong sequence similarity is plagued with various technical problems, which encompass over half of representative domain pairs and make the examination a tedious task. These problems arise at all stages, from experiment (genetic construct, structure determination) to data processing (generating PDB file and SCOP domain) and data analysis (profile, alignment, structure superposition). On the other hand, careful investigation reveals interesting examples of homologs with distinct structures and advances our understanding of protein evolution. We see that insertions, extensions, and duplications decorate and expand evolutionary core; deletions reduce the core, sometimes beyond recognition, potentially resulting in reorientation of structural elements. Topology and mutual arrangement of secondary structures may change due to circular permutation or domain swapping. Finally, combination of several such events makes for the largest structural differences between homologs.
Evolution of function and active sites up
Proteins are quite promiscuous in ways they change functions in evolution. Some enzymes lose active sites and become binding proteins, while binding modules gain active sites and become enzymes. A few examples are mentioned here.
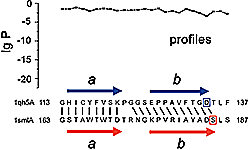
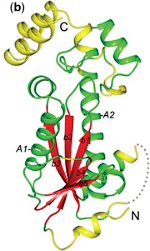
Many examples of enzymes that have lost their catalytic activity and perform other biological functions are known. The opposite situation is rare. A previously unnoticed structural similarity between the λ integrase family (Int) proteins and the AraC family of transcriptional activators implies that the Int family evolved by duplication of an ancient DNA-binding homeodomain-like module, which acquired enzymatic activity. The two helix-turn-helix (HTH) motifs in Int proteins incorporate catalytic residues and participate in DNA binding. The active site of Int proteins, which include the type IB topoisomerases, is formed at the domain interface and the catalytic tyrosine residue is located in the second helix of the C-terminal HTH motif. Structural analysis of other 'tyrosine' DNA-breaking/rejoining enzymes with similar enzyme mechanisms, namely prokaryotic topoisomerase I, topoisomerase II and archaeal topoisomerase VI, reveals that the catalytic tyrosine is placed in a HTH domain as well. Surprisingly, the location of this tyrosine residue in the structure is not conserved, suggesting independent, parallel evolution leading to the same catalytic function by homologous HTH domains. The 'tyrosine' recombinases give a rare example of enzymes that evolved from ancient DNA-binding modules and present a unique case for homologous enzymatic domains with similar catalytic mechanisms but different locations of catalytic residues, which are placed at non-homologous sites. PDF
Comparisons of serine/threonine protein kinase (PK) and type IIβ phosphatidylinositol phosphate kinase (PIPK) structures with each other and also with other proteins reveal structural and functional similarity between the two kinases and proteins of the glutathione synthase fold (ATP-grasp). This suggests that these enzymes are evolutionarily related. The structure of PIPK, which clearly resembles both PK and ATP-grasp, provides a link between the two proteins and establishes that the C-terminal domains of PK, PIPK and ATP-grasp share the same fold. It is likely that protein kinases evolved from metabolic enzymes with ATP-grasp fold through lipid PIPK-like kinases. PDF
Zn-dependent carboxypeptidases (ZnCP) cleave off the C-terminal amino acid residues from proteins and peptides. We analyzed a superfamily that unites classical ZnCP with other enzymes, most of which are known (or likely) to participate in metal-dependent peptide bond cleavage, but not necessarily in polypeptide substrates. It is demonstrated that aspartoacylase (ASP gene) and succinylglutamate desuccinylase (ASTE gene) are members of the ZnCP family. The Zn-binding site along with the structural core of the protein is shown to be conserved between ZnCP and another large family of hydrolases that includes mostly aminopeptidases (ZnAP). Both families (ZnCP and ZnAP) include not only proteases but also enzymes that perform N-deacylation, and enzymes that catalyze N-desuccinylation of amino acids. This is a result of functional convergence that apparently occurred after the divergence of the two families. PDF
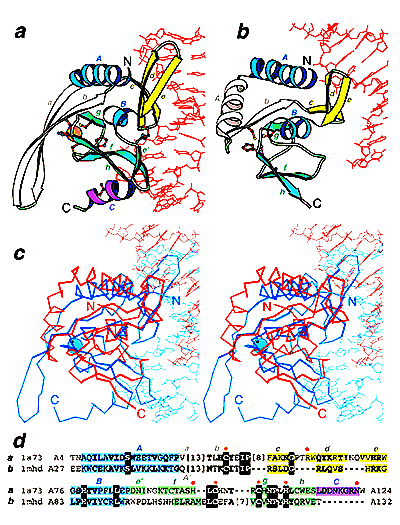
Helix-hairpin-helix (HhH) is a widespread motif involved in non-sequence-specific DNA binding. The majority of HhH motifs function as DNA-binding modules, however, some of them are used to mediate protein-protein interactions or have acquired enzymatic activity by incorporating catalytic residues (DNA glycosylases). From sequence and structural analysis of HhH-containing proteins we conclude that most HhH motifs are integrated as a part of a five-helical domain, termed (HhH)2 domain here. It typically consists of two consecutive HhH motifs that are linked by a connector helix and displays pseudo-2-fold symmetry. (HhH)2 domains show clear structural integrity and a conserved hydrophobic core composed of seven residues, one residue from each alpha-helix and each hairpin, and deserves recognition as a distinct protein fold. In addition to known HhH in the structures of RuvA, RadA, MutY and DNA-polymerases, we have detected new HhH motifs in sterile alpha motif and barrier-to-autointegration factor domains, the alpha-subunit of Escherichia coli RNA-polymerase, DNA-helicase PcrA and DNA glycosylases. Statistically significant sequence similarity of HhH motifs and pronounced structural conservation argue for homology between (HhH)2 domains in different protein families. Our analysis helps to clarify how non-symmetric protein motifs bind to the double helix of DNA through the formation of a pseudo-2-fold symmetric (HhH)2 functional unit. PDF
Smad proteins are eukarytic transcription regulators in the TGF-beta signaling cascade. Using a combination of sequence and structure-based analyses, we argue that MH1 domain of Smad is homologous to the diverse His-Me finger endonuclease family enzymes. The similarity is particularly extensive with the I-PpoI endonuclease. In addition to the global fold similarities, both proteins possess a conserved motif of three cysteine residues and one histidine residue which form a zinc-binding site in I-PpoI. Sequence and structure conservation in the motif region strongly suggest that MH1 domain may also incorporate a metal ion in its structural core. This was later verified experimentally. MH1 of Smad3 and I-PpoI exhibit similar nucleic acid binding mode and interact with DNA major groove through an antiparallel beta-sheet. MH1 is an example of transcription regulator derived from the ancient enzymatic domain that lost its catalytic activity but retained DNA-binding sites. PDF1 PDF2
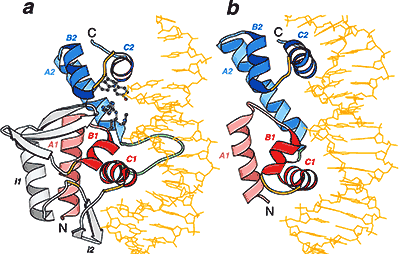
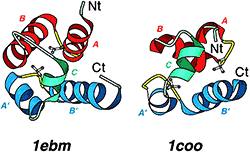
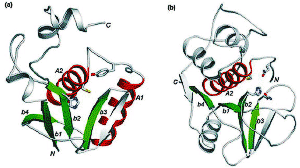
Detection of similarity is particularly difficult for small proteins and thus connections between many of them remain unnoticed. Structure and sequence analysis of several metal-binding proteins reveals unexpected similarities in structural domains classified as different protein folds in SCOP and suggests unification of seven folds that belong to two protein classes. The common motif that we termed treble clef finger, forms the protein structural core and is 25-45 residues long. The treble clef motif is assembled around the central zinc ion and consists of a zinc knuckle, loop, β-hairpin and an α-helix. The knuckle and the first turn of the helix each incorporate two zinc ligands. Treble clef domains constitute the core of many structures such as ribosomal proteins L24E and S14, RING fingers, protein kinase cysteine-rich domains, nuclear receptor-like fingers, LIM domains, phosphatidylinositol-3-phosphate-binding domains and His-Me finger endonucleases. The treble clef finger is a uniquely versatile motif adaptable for various functions. This small domain with a 25 residue structural core can accommodate eight different metal-binding sites and can have many types of functions from binding of nucleic acids, proteins and small molecules, to catalysis of phosphodiester bond hydrolysis. Treble clef motifs are frequently incorporated in larger structures or occur in doublets. Our analysis suggests that the treble clef motif defines a distinct structural fold found in proteins with diverse functional properties and forms one of the major zinc finger groups. PDF
Variability of evolutionary rates between sites and proteins up
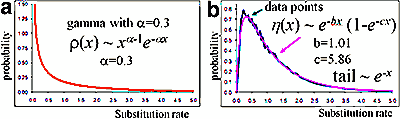
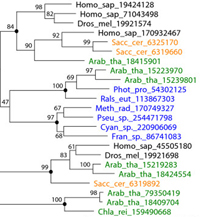
Accumulation of complete genome sequences of diverse organisms creates new possibilities for evolutionary inferences from whole-genome comparisons. We analyzed the distributions of substitution rates among proteins encoded in 19 complete genomes (the interprotein rate distribution). To estimate these rates, it is necessary to employ another fundamental distribution, that of the substitution rates among sites in proteins (the intraprotein distribution). Using two independent approaches, we show that intraprotein substitution rate variability appears to be significantly greater than generally accepted. This yields more realistic estimates of evolutionary distances from amino-acid sequences, which is critical for evolutionary-tree construction. We demonstrate that the interprotein rate distributions inferred from the genome-to-genome comparisons are similar to each other and can be approximated by a single distribution with a long exponential shoulder. This suggests that a generalized version of the molecular clock hypothesis may be valid on genome scale. We also use the scaling parameter of the obtained interprotein rate distribution to construct a rooted whole-genome phylogeny. The topology of the resulting tree is largely compatible with those of global rRNA-based trees and trees produced by other approaches to genome-wide comparison. PDF
Mathematical modeling of sequence and structure evolution up
We proposed a general model for estimating the number of amino acid substitutions per site (d) from the fraction of identical residues between two sequences (q). The well-known Poisson-correction formula q = exp(-d) corresponds to a site-independent and amino-acid-independent substitution rate. Equation q = (1 - exp(-2d))/2d, derived for the case of substitution rates that are site-independent, but vary among amino acids, approximates closely the empirical method, suggested by Dayhoff et al. (1978). Equation q = 1/(1 + d) describes the case of substitution rates that are amino acid-independent but vary among sites. Lastly, equation q = [ln(1 + 2d)]/2d accounts for the general case where substitution rates can differ for both amino acids and sites. PDF
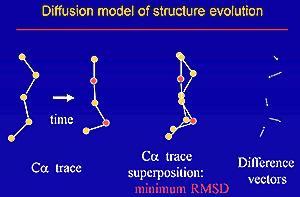
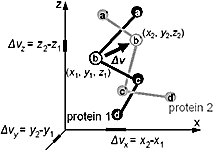
We derived new equations to estimate the number of amino acid substitutions per site between two homologous proteins from the root mean square (RMS) deviation between two spatial structures and from the fraction of identical residues between two sequences. The equations are based on evolutionary models, analyzing predominantly structural changes and not sequence changes. Evolution of spatial structure is treated as a diffusion in an elastic force field. Diffusion accounts for structural changes caused by amino acid substitutions, and elastic force reflects selection, which preserves protein fold. Obtained equations are supported by analysis of protein spatial structures. PDF
Simulation of sequence and structure evolution up
The biological function of a protein often depends on the formation of an ordered structure in order to support a smaller, chemically active configuration of amino acids against thermal fluctuations. We explore the development of proteins evolving to satisfy this requirement using an off-lattice polymer model in which monomers interact as low resolution amino acids. To evolve the model, we construct a Markov process in which sequences are subjected to random replacements, insertions, and deletions and are selected to recover a predefined minimum number of solid-ordered monomers using the Lindemann melting criterion. We show that polymers generated by this process consistently fold into soluble, ordered globules of similar length and complexity to small protein motifs. To compare the evolution of the globules with proteins, we analyze the statistics of amino acid replacements, the dependence of site mutation rates on solvent exposure, and the dependence of structural distance on sequence distance for homologous alignments. Despite the simplicity of the model, the results display a surprisingly close correspondence with protein data. PDF
Definition of protein domains up
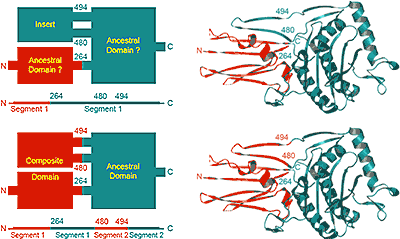
Proteins are composed of domains. Domains are usually defined as globular units in protein structures. Domains are somewhat separate from each other spatially and can recombine with each other in evolution to form various proteins. Each domain frequently carries out its own function, or the functional site may be formed at the domain interface. Analysis of domains is essential for understanding of proteins. However, while everyone agrees on the domain importance, opinions differ greatly about the criteria for domain definition, and existing software tools are inconsistent with each other in domain parse. Researchers think about domains from the position of structural compactness, sequence similarity and continuity, evolutionary origin, folding or function. Different criteria lead to different domain definitions. Nevertheless, our experience with protein sequence-structural analysis indicates that it might be possible to bring these criteria together for a biologically reasonable domain parse. We applied our conceptual view on protein domains to the most challenging group of proteins currently defined as "multidomain" class in SCOP. These proteins are large and are composed of several frequently intertwined domains, making domain definition particularly challenging. For the first time, domain definitions for these proteins are provided and can be used to train domain-parsing software, or to study evolution of these proteins. PDF
Intrinsic structural disorder in proteins up
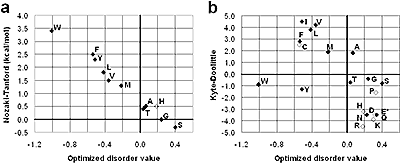
X-ray crystallographic protein structures often contain disordered regions that are observed as missing electron density. Diffraction data may give little or no direct evidence as to the specific nature of disordered regions. We have developed a weighted window-based disorder predictor optimized using crystallographic data. Performance of a predictor is strongly influenced by chain termini. Optimized score adjustment values for amino- and carboxy-terminal positions demonstrate a simple, monotonic relationship between disorder and residue distance from termini. This optimized disorder predictor performs similarly to DISOPRED2 on crystallographically disordered regions. Data-optimized residue disorder propensities show strong linear correlation with experimentally determined amino acid transfer energies between water and hydrogen-bonding organic solvents, which primarily reflect residue hydrophobicity (exemplified by the Nozaki–Tanford hydrophobicity scale). Disorder propensities do not correlate as well with transfer energies between water and apolar solvents, which primarily reflect a different hydropathic property: residue hydrophilicity (also reflected by the Kyte-Doolittle hydropathy scale). Our results suggest that while hydrophobic side-chain interactions are primarily involved in determining stability of the folded conformation, hydrogen bonding, and similar polar interactions are primarily involved in conformational and interaction specificity. PDF
Non-randomness of protein structure topologies up
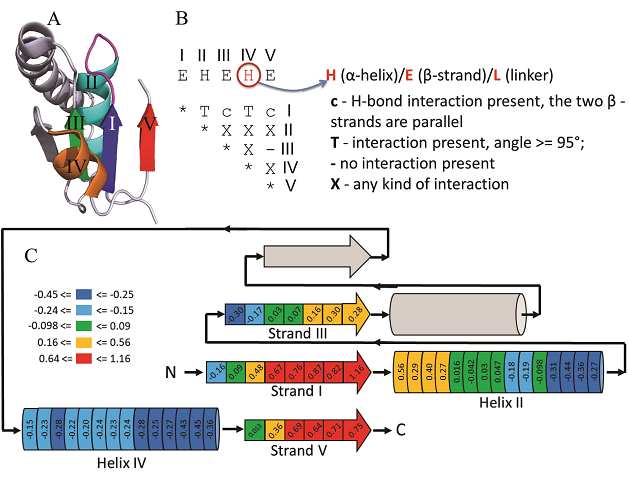
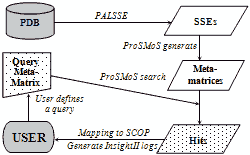
Most protein structures are folded as semi-regular arrays of parallel and anit-parallel secondary structural elements. It is possible to approximate locations of secondary structural elements in such structures as points on a hexagonal 2D grid. The resulting topology diagram is a succinct description of a protein fold. Interactions between neighboring secondary structures and handedness of connections between triplets of secondary structures uniquely determine the correspondence between the topology diagram and real protein structure. We exhaustively enumerate topology diagrams for small number of secondary structures (3 to 8) and find proteins that contain these as substructures. Each diagram is converted into a ProSMoS meta-matrix, and PDB database is searched with it. The results show highly non-random distribution of real structures by topology types, and symmetric, simple and regular topologies are more abundant in proteins. 3D and topology diagrams of several common protein folds can be found here.
Dependence of folding on structure topology up
We consider a nonstatistical, computationally fast experiment to identify important topological constraints in folding small globular proteins of about 100-200 amino acids. In this experiment, proteins are expanded mechanically along a path of steepest increase in the free space around residues. The pathways are often consistent with folding scenarios reported in kinetics experiments and most accurately describe obligatory or mechanic folding proteins. The results suggest that certain topological "defects" in proteins lead to preferred, entropically favorable channels down their free energy landscapes. PDF
We studied a nucleation-growth model of protein folding and extend it to describe larger proteins with multiple folding units. The model is of one of an extremely simple type in which amino acids are allowed just two states – either folded (frozen) or unfolded. Its energetics are heterogeneous and Gō-like, the energy being defined in terms of the number of atom-to-atom contacts that would occur between frozen amino acids in the native crystal structure of the protein. Each collective state of the amino acids is intended to represent a small free energy microensemble consisting of the possible configurations of unfolded loops, open segments, and free ends constrained by the cross-links that form between folded parts of the molecule. We approximate protein free energy landscapes by an infinite subset of these microensemble topologies in which loops and open unfolded segments can be viewed roughly as independent objects for the purpose of calculating their entropy, and we develop a means to implement this approximation in Monte Carlo simulations. We show that this approach describes transition state structures (φ–values) more accurately and identifies folding intermediates that were unavailable to previous versions of the model that restricted the number of loops and nuclei. PDF
A recent study of experimental results for flavodoxin-like folds suggests that proteins from this family may exhibit a similar, signature pattern of folding intermediates. We study the folding landscapes of three proteins from the flavodoxin family (CheY, apoflavodoxin, and cutinase) using a simple nucleation and growth model that accurately describes both experimental and simulation results for the transition state structure, and the structure of on-pathway and misfolded intermediates for CheY. Although the landscape features of these proteins agree in basic ways with the results of the study, the simulations exhibit a range of folding behaviors consistent with two alternate folding routes corresponding to nucleation and growth from either side of the central β-strand. PDF
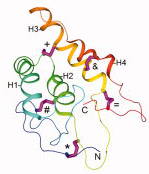
The B domain of staphylococcal protein A (BdpA) is a small helical protein that has been studied intensively in kinetics experiments and detailed computer simulations that include explicit water. The simulations indicate that BdpA needs to reorganize in crossing the transition barrier to facilitate folding its C-terminal helix (H3) onto the nucleus formed from helices H1 and H2. This process suggests frustration between two partially ordered forms of the protein, but recent φ–value measurements indicate that the transition structure is relatively constant over a broad range of temperatures. Here we develop a simplistic model to investigate the folding transition in which properties of the free energy landscape can be quantitatively compared with experimental data. The model is a continuation of the Muñoz-Eaton model to include the intermittency of contacts between structured parts of the protein, and the results compare variations in the landscape with denaturant and temperature to φ–value measurements and chevron plots of the kinetic rates. The topography of the model landscape (in particular, the feature of frustration) is consistent with detailed simulations even though variations in the φ–values are close to measured values. The transition barrier is smaller than indicated by the chevron data, but it agrees in order of magnitude with a similar alpha-carbon type of model. Discrepancies with the chevron plots are investigated from the point of view of solvent effects, and an approach is suggested to account for solvent participation in the model. PDF
Interpretation of clinically important mutations up
We are collaborating with several research groups to understand the molecular effect of disease-causing mutations. Four examples, all from the Hobbs lab, are given here.
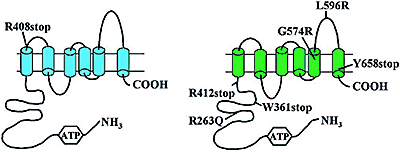
In healthy individuals, acute changes in cholesterol intake produce modest changes in plasma cholesterol levels. A striking exception occurs in sitosterolemia, an autosomal recessive disorder characterized by increased intestinal absorption and decreased biliary excretion of dietary sterols, hypercholesterolemia, and premature coronary atherosclerosis. We identified seven different mutations in two adjacent, oppositely oriented genes that encode new members of the adenosine triphosphate (ATP)-binding cassette ABC transporter family (six mutations in ABCG8 and one in ABCG5) in nine patients with sitosterolemia. The two genes are expressed at highest levels in liver and intestine and, in mice, cholesterol feeding up-regulates expressions of both genes. These data suggest that ABCG5 and ABCG8 normally cooperate to limit intestinal absorption and to promote biliary excretion of sterols, and that mutated forms of these transporters predispose to sterol accumulation and atherosclerosis. PDF
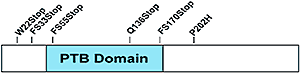
Atherogenic low density lipoproteins are cleared from the circulation by hepatic low density lipoprotein receptors (LDLR). Two inherited forms of hypercholesterolemia result from loss of LDLR activity: autosomal dominant familial hypercholesterolemia (FH), caused by mutations in the LDLR gene, and autosomal recessive hypercholesterolemia (ARH), of unknown etiology. Here we map the ARH locus to an approximately 1-centimorgan interval on chromosome 1p35 and identify six mutations in a gene encoding a putative adaptor protein (ARH). ARH contains a phosphotyrosine binding (PTB) domain, which in other proteins binds NPXY motifs in the cytoplasmic tails of cell-surface receptors, including the LDLR. ARH appears to have a tissue-specific role in LDLR function, as it is required in liver but not in fibroblasts. PDF
Elevated levels of circulating low-density lipoprotein cholesterol (LDL-C) play a central role in the development of atherosclerosis. Mutations in proprotein convertase subtilisin/kexin type 9 (PCSK9) that are associated with lower plasma levels of LDL-C confer protection from coronary heart disease. Here, we show that four severe loss-of-function mutations prevent the secretion of PCSK9 by disrupting synthesis or trafficking of the protein. In contrast to recombinant wild-type PCSK9, which was secreted from cells into the medium within 2 hours, the severe loss-of-function mutations in PCSK9 largely abolished PCSK9 secretion. This finding predicted that circulating levels of PCSK9 would be lower in individuals with the loss-of-function mutations. Immunoprecipitation and immunoblotting of plasma for PCSK9 provided direct evidence that the serine protease is present in the circulation and identified the first known individual who has no immunodetectable circulating PCSK9. This healthy, fertile college graduate, who was a compound heterozygote for two inactivating mutations in PCSK9, had a strikingly low plasma level of LDL-C (14 mg/dL). The very low plasma level of LDL-C and apparent good health of this individual demonstrate that PCSK9 plays a major role in determining plasma levels of LDL-C and provides an attractive target for LDL-lowering therapy. PDF
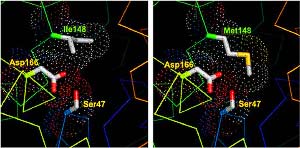
Obesity and insulin resistance are associated with deposition of triglycerides in tissues other than adipose tissue. Previously, we showed that a missense mutation (I148M) in PNPLA3 (patatin-like phospholipase domain-containing 3 protein) is associated with increased hepatic triglyceride content in humans. Here we examined the effect of the I148M substitution on the enzymatic activity and cellular location of PNPLA3. Structural modeling predicted that the substitution of methionine for isoleucine at residue 148 would restrict access of substrate to the catalytic serine. In vitro assays using recombinant PNPLA3 partially purified from Sf9 cells confirmed that the wild type enzyme hydrolyzes emulsified triglyceride and that the I148M substitution abolishes this activity. Expression of PNPLA3-I148M, but not wild type PNPLA3, in cultured hepatocytes or in the livers of mice increased cellular triglyceride content. Cell fractionation studies revealed that approximately 90% of wild type PNPLA3 partitioned between membranes and lipid droplets; substitution of isoleucine for methionine at position 148 did not alter the subcellular distribution of the protein. These data are consistent with PNPLA3-I148M promoting triglyceride accumulation by limiting triglyceride hydrolysis. PDF
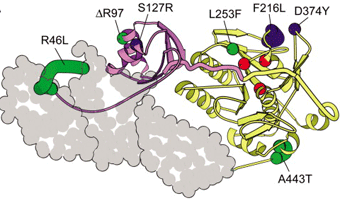
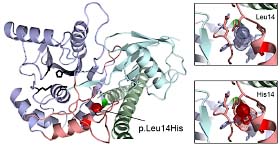
Mapping disease causing missense mutations that exist in protein domains with known structure can shed insight into the alteration of protein function and its impact on disease. We have collaborated with the Brugarolas lab to interpret such missense mutations that occur in renal cell carcinoma (RCC). The nuclear deubiquitinase BAP1 is inactivated in 15% of clear cell RCC and a novel germline mutation in the gene predisposes to familial RCC. We constructed a BAP1 structure model based on related Ubiquitin C-terminal Hydrolase (UCH) family members Uch-L3 and Uch37 to understand missense mutations that map to the catalytic UCH domain. Two of the mutations that did not abrogate BAP1 expression (p.G13V and p.P170L) disrupted side chains implicated in either an intramolecular interaction with the ULD domain (Gly13) or ubiquitin binding (Phe170), and highlight the importance of these interactions for tumor suppressor function (PDF1 ). The same model was used to interpret the BAP1 germline mutation (p.L14H) that predisposes to familial RCC. Leucine 14 maps to the first helix of the UCH domain and is physically adjacent to two previously identified pathogenic RCC mutations. The altered position helps organize a crossover loop and other flexible portions of the UCH domain that order upon ubiquitin binding, and forms a portion of the interaction surface for the ULD tail. Mutation of this residue to histidine is predicted to increase the effective volume of the side chain, possibly causing steric clashes with surrounding residues, and may prevent productive ubiquitin binding (PDF2 ).
Single-amino acid variations (SAVs) (single-nucleotide changes that alter amino acids) in protein-coding regions are one of the major causes of human phenotypic variation and diseases. These are routinely found in whole genome and exome sequencing. Evaluating the functional impact of such genomic alterations is crucial for diagnosis of genetic disorders. We developed DeepSAV, a deep-learning convolutional neural network to differentiate disease-causing and benign SAVs based on a variety of protein sequence, structural and functional properties. Our method outperforms most stand-alone programs and has similar predictive power as some of the best available. We transformed DeepSAV scores of rare SAVs observed in the general population into a mutation severity measure of protein-coding genes. This measure reflects a gene's tolerance to deleterious missense mutations and serves as a useful tool to study gene-disease associations. Genes implicated in cancer, autism, and viral interaction are found by this measure as intolerant to mutations, while genes associated with a number of other diseases are scored as tolerant. Among known disease-associated genes, those that are mutation-intolerant are likely to function in development and signal transduction pathways, while those that are mutation-tolerant tend to encode metabolic proteins and proteins targeted to mitochondria, such as mitochondrial ribosomal proteins (PDF ).
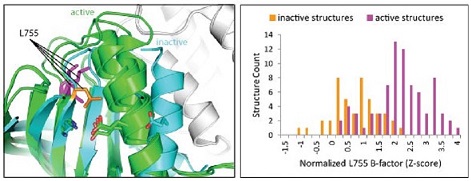
The EGFR-like protein kinase human epidermal growth factor receptor 2 (ERBB2/HER2) is frequently activated in breast cancers, with 2% to 4% having HER2 missense mutations. By comparing 122 EGFR-like kinase structures in active and inactive conformations, we helped identify the mechanism of activation for the missense mutation L755S. The residue L755 forms a stable hydrophobic core in inactive conformations, while it becomes flexible in active conformations. This flexibility is revealed in a distribution of normalized B-factors for L755 shifting to higher values in active conformations. The mutation to Ser promotes this flexibility and thus activates HER2. (PDF )
Search for proteins responsible for new activities up
What can be more exciting than discovery of molecules with novel activities! Computational analysis may point to the right candidates.
In collaboration with the Brown and Goldstein Lab, we identified acyltransferase that octanoylates ghrelin. Ghrelin is a 28 amino acid, appetite-stimulating peptide hormone secreted by the food-deprived stomach. Serine-3 of ghrelin is acylated with an eight-carbon fatty acid, octanoate, which is required for its endocrine actions. Here, we identify GOAT (Ghrelin O-Acyltransferase), a polytopic membrane-bound enzyme that attaches octanoate to serine-3 of ghrelin. Analysis of the mouse genome revealed that GOAT belongs to a family of 16 hydrophobic membrane-bound acyltransferases that includes Porcupine, which attaches long-chain fatty acids to Wnt proteins. GOAT is the only member of this family that octanoylates ghrelin when coexpressed in cultured endocrine cell lines with prepro-ghrelin. GOAT activity requires catalytic asparagine and histidine residues that are conserved in this family. Consistent with its function, GOAT mRNA is largely restricted to stomach and intestine, the major ghrelin-secreting tissues. Identification of GOAT will facilitate the search for inhibitors that reduce appetite and diminish obesity in humans. PDF
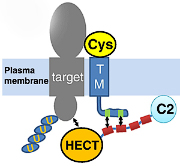
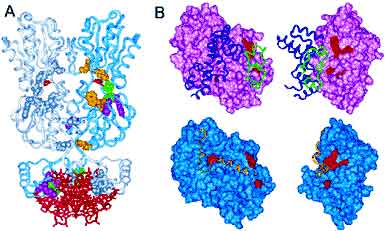
In collaboration with the Orth Lab, we show that AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. The Vibrio parahaemolyticus type III effector VopS is implicated in cell rounding and the collapse of the actin cytoskeleton by inhibiting Rho GTPases. We found that VopS (from the FIC domain superfamily) could act as an AMPylator to covalently modify a conserved threonine residue on Rho, Rac, and Cdc42 with adenosine 5'-monophosphate. The resulting AMPylation prevented interaction of Rho GTPases with downstream effectors, thereby inhibiting actin assembly in the infected cell. Eukaryotic proteins were also directly modified with AMP, potentially expanding the repertoire of posttranslational modifications for molecular signaling. This is the first functional characterization of the FIC domain: a larger universal family of formely hypothetical proteins with a few structures determined by structural genomics initiatives. PDF
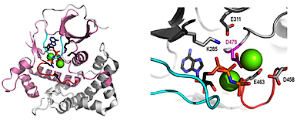
In collaboration with the Bruick Lab, we characterized iron binding protein FBXL5. Cellular iron homeostasis is maintained by the coordinate posttranscriptional regulation of genes responsible for iron uptake, release, use, and storage through the actions of the iron regulatory proteins IRP1 and IRP2. However, the manner in which iron levels are sensed to affect IRP2 activity is poorly understood. We found that an E3 ubiquitin ligase complex containing the FBXL5 protein targets IRP2 for proteasomal degradation. The stability of FBXL5 itself was regulated, accumulating under iron- and oxygen-replete conditions and degraded upon iron depletion. We identified a hemerythrin-like domain at the N-terminus of FBXL5 that binds iron and oxygen, acting as a ligand-dependent regulatory switch mediating FBXL5’s differential stability. Residues 1 to 161 of the human FBXL5 protein are predicted to contain five α-helices encompassing several conserved histidine and glutamic acid residues, similar to hemerythrin-like four-helix up and down bundles with an additional C-terminal helix packed against the core. Although not previously reported in mammalian proteins, hemerythrin domains have been frequently reported to contain m-oxo diiron centers that reversibly bind oxygen and often function as O2- transport proteins, O2 sensors, or metal storage depots in marine invertebrates and bacteria. These observations suggest a mechanistic link between iron sensing via the FBXL5 hemerythrin domain, IRP2 regulation, and cellular responses to maintain mammalian iron homeostasis. PDF
In collaboration with the Liu Lab, we discovered a novel type of endoribonuclease. The catalytic engine of RNA interference (RNAi) is the RNA-induced silencing complex (RISC), wherein the endoribonuclease Argonaute and single-stranded small interfering RNA (siRNA) direct target mRNA cleavage. We reconstituted long double-stranded RNA- and duplex siRNA-initiated RISC activities with the use of recombinant Drosophila Dicer-2, R2D2, and Ago2 proteins. We used this core reconstitution system to purify an RNAi regulator that we term C3PO (component 3 promoter of RISC), a complex of Translin and Trax. C3PO is a Mg2+-dependent endoribonuclease that promotes RISC activation by removing siRNA passenger strand cleavage products. We identified residues corresponding to the endonuclease active site by mapping the C3PO sequence to the human translin structure. To identify these residues, we performed a multisequence alignment of Translin and Trax and observed three acidic residues (Glu123, Glu126, and Asp204) that were invariant in Trax but missing in Translin. Furthermore, modeling the structure of Drosophila Trax after the crystal structure of human Translin revealed that these residues existed in close spatial proximity, which suggests that they may coordinate Mg2+ for catalysis. These studies establish an in vitro RNAi reconstitution system and identify C3PO as a key activator of the core RNAi machinery. PDF
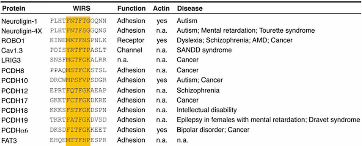
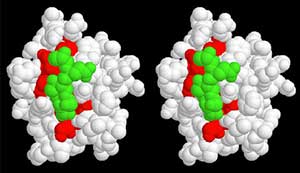
In collaboration with the Rosen Lab, we discovered 115 potential WIRS motif containing proteins that may bind to the WAVE regulatory complex (WRC). Membrane or membrane-associated proteins in Swiss-Prot database were queried with the motif pattern, Φ-F-x-T/S-F-X-X (Φ for bulky hydrophobic residues), together with secondary structure features to mimic the binding mode seen in the structure. Many of these proteins are enriched in the nervous or immune systems, although others are widely expressed. Of these, only five had been previously shown to interact with the WRC biochemically or genetically. Furthermore, only a small number had been previously connected with the actin cytoskeleton. GST-fused cytoplasmic tails of 18 of these potential ligands were tested using pull-down assays. We found that 13 of them bound the WT WRC, but not a mutant whose WIRS-binding surface was disrupted. These diverse WIRS-containing tails also have various effects on WRC activity. We have identified a consensus peptide motif, WIRS, which specifically binds to a unique surface of WRC and characterized a large family of potential WRC ligands unique to metazoans. PDF
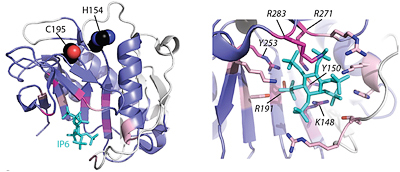
Vibrio parahaemolyticus is a Gram-negative halophilic bacterium and one of the leading causes of food-borne gastroenteritis. Its genome harbors both Type III Secretion Systems (T3SS) and Type VI Secretion Systems (T6SS) that deliver virulence effector proteins into target cells. We have collaborated with the Orth Lab to shed light on the function of several of these effectors. We helped identify a conserved bacterial phosphoinositide-binding domain (BPD) that is found in functionally diverse T3SS effectors of both plant and animal pathogens that delivers effectors specifically to the inner membrane of host cells (PDF1 ). Using comparative proteomics, we identified two previously unidentified T6SS effectors that contained a conserved motif. Our bioinformatics analyses revealed that this N-terminal motif, named MIX (marker for type six effectors), is found in numerous polymorphic bacterial proteins that are primarily located in the T6SS genome neighborhood. Several of the MIX-containing proteins functioned as effectors that killed neighboring bacterial cells. Thus, our findings identified numerous uncharacterized T6SS effectors that can lead to the discovery of new biological mechanisms of bacterial warfare (PDF2 ). The Orth lab also identified a T3SS2 effector protein (VPA1380) that is toxic when expressed in yeast. Our bioinformatics analyses revealed that VPA1380 is similar to the inositol hexakisphosphate (IP6)-inducible cysteine protease domains of several large bacterial toxins. Structure modeling, combined with sequence conservation analysis suggested mutations in conserved catalytic residues and residues in the putative IP6-binding pocket that abolished toxicity in yeast. Furthermore, VPA1380 was not toxic in IP6 deficient yeast cells. Therefore, our findings suggest that VPA1380 is a cysteine protease that requires IP6 as an activator (PDF3 ).
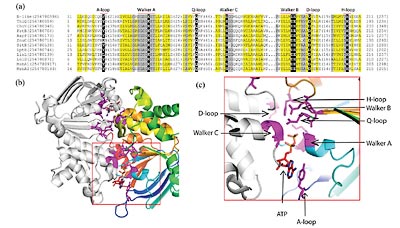
Protein kinases constitute one of the largest and functionally diverse gene families, with members representing almost 2% of the human genome. The protein kinase catalytic domain is conserved in sequence and numerous structures have been solved, revealing an active site at the interface of two lobes. Conserved sequence features that contribute to the active site include a glycine-containing loop and an ion pair in the N-terminal lobe, as well as an ion coordinating aspartic acid and a catalytic aspartic acid in the C-terminal lobe. In collaboration with the Dixon Lab, using these features and sensitive profile-based sequence detection methods, we identified a group of secreted kinases that are distantly related to the protein kinase-like superfamily. One of these kinases, Fam20C, is the physiological casein kinase and phosphorylates a diverse array of secreted substrates (PDF1 ). We have also used knowledge of the catalytic protein kinase structure/function to help understand the mechanism of cancer causing mutations in the protein kinase MET (PDF2 ), to help understand drug sensitivity and resistance in tumors (PDF3 ), and to help identify activation mechanism of JAK2 (PDF4 ).
We extended our prediction of novel protein kinase like domains to include SelO, which was first thought to be lacking the catalytic Asp. However, the binding site of ATP has flipped in the SelO structure, making the kinase an AMPylator that transfers AMP from ATP to protein substrates. Sequence alignments suggested the catalytic Asp migrated to SelO residue D252 and pointed to the role of a disulfide bond regulating activity. (PDF5 )
Sequence-structure-function relationship in protein families up
We spend quite a bit of time analyzing individual protein families with the goal to further our understanding of their evolution, structure and function. Several semi-randomly selected examples of our work are given here.
Nitrogen regulatory (PII) proteins are signal transduction molecules involved in controlling nitrogen metabolism in prokaryots. PII proteins integrate the signals of intracellular nitrogen and carbon status into the control of enzymes involved in nitrogen assimilation. Using elaborate sequence similarity detection schemes, we show that five clusters of orthologs (COGs) and several small divergent protein groups belong to the PII superfamily and predict their structure to be a (βαβ)2 ferredoxin-like fold. Proteins from the newly emerged PII superfamily are present in all major phylogenetic lineages. The PII homologs are quite diverse, with below random (as low as 1%) pairwise sequence identities between some members of distant groups. Despite this sequence diversity, evidence suggests that the different subfamilies retain the PII trimeric structure important for ligand-binding site formation and maintain a conservation of conservations at residue positions important for PII function. Because most of the orthologous groups within the PII superfamily are composed entirely of hypothetical proteins, our remote homology-based structure prediction provides the only information about them. Analogous to structural genomics efforts, such prediction gives clues to the biological roles of these proteins and allows us to hypothesize about locations of functional sites on model structures or rationalize about available experimental information. For instance, conserved residues in one of the families map in close proximity to each other on PII structure, allowing for a possible metal-binding site in the proteins coded by the locus known to affect sensitivity to divalent metal ions. Presented analysis pushes the limits of sequence similarity searches and exemplifies one of the extreme cases of reliable sequence-based structure prediction. In conjunction with structural genomics efforts to shed light on protein function, our strategies make it possible to detect homology between highly diverse sequences and are aimed at understanding the most remote evolutionary connections in the protein world. PDF
The O-linked GlcNAc transferases (OGTs) are a recently characterized group of largely eukaryotic enzymes that add a single beta-N-acetylglucosamine moiety to specific serine or threonine hydroxyls. In humans, this process may be part of a sugar regulation mechanism or cellular signaling pathway that is involved in many important diseases, such as diabetes, cancer, and neurodegeneration. However, no structural information about the human OGT exists, except for the identification of tetratricopeptide repeats (TPR) at the N terminus. The locations of substrate binding sites are unknown and the structural basis for this enzyme's function is not clear. Here, remote homology is reported between the OGTs and a large group of diverse sugar processing enzymes, including proteins with known structure such as glycogen phosphorylase, UDP-GlcNAc 2-epimerase, and the glycosyl transferase MurG. This relationship, in conjunction with amino acid similarity spanning the entire length of the sequence, implies that the fold of the human OGT consists of two Rossmann-like domains C-terminal to the TPR region. A conserved motif in the second Rossmann domain points to the UDP-GlcNAc donor binding site. This conclusion is supported by a combination of statistically significant PSI-BLAST hits, consensus secondary structure predictions, and a fold recognition hit to MurG. Additionally, iterative PSI-BLAST database searches reveal that proteins homologous to the OGTs form a large and diverse superfamily that is termed GPGTF (glycogen phosphorylase/glycosyl transferase). Up to one-third of the 51 functional families in the CAZY database, a glycosyl transferase classification scheme based on catalytic residue and sequence homology considerations, can be unified through this common predicted fold. GPGTF homologs constitute a substantial fraction of known proteins: 0.4% of all non-redundant sequences and about 1% of proteins in the Escherichia coli genome are found to belong to the GPGTF superfamily. PDF
Sec61p/SecYEG complexes mediate protein translocation across membranes and are present in both eukaryotes and bacteria. Whereas homologues of Sec61alpha/SecY and Sec61gamma/SecE exist in archaea, identification of the third component (Sec61beta or SecG) has remained elusive. Using PSI-BLAST, the archaeal counterpart of Sec61beta has been detected. With the identification of the Sec61beta motif, functions for a universal family of archaeal proteins can be predicted and the archaeal translocon system can be definitively detected. PDF
Sequence and structure – based searching strategies have proven useful in the identification of remote homologs and have facilitated both structural and functional predictions of many uncharacterized protein families. We implement these strategies to predict the structure of and to classify a previously uncharacterized cluster of orthologs (COG3019) in the thioredoxin-like fold superfamily. The results of each searching method indicate that thioltransferases are the closest structural family to COG3019. We substantiate this conclusion using the ab initio structure prediction method ROSETTA, which generates a thioredoxin-like fold similar to that of the glutaredoxin-like thioltransferase (NrdH) for a COG3019 target sequence. This structural model contains the thiol-redox functional motif CYS-X-X-CYS in close proximity to other absolutely conserved COG3019 residues, defining a novel thioredoxin-like active site that potentially binds metal ions. Finally, the rosetta-derived model structure assists us in assembling a global multiple-sequence alignment of COG3019 with two other thioredoxin-like fold families, the thioltransferases and the bacterial arsenate reductases (ArsC). PDF
Using a recently developed program (SCOPmap) designed to automatically assign new protein structures to existing evolutionary-based classification schemes, we identify a evolutionarily conserved domain (EDD) common to three different folds: mannose transporter EIIA domain (EIIA-man), dihydroxyacetone kinase (Dak), and DegV. Several lines of evidence support unification of these three folds into a single superfamily: statistically significant sequence similarity detected by PSI-BLAST; "closed structural grouping" using DALI Z-scores (each protein inside a group finds all other group members with scores higher than those to proteins outside the group) that includes only these proteins sharing a unique alpha-helical hairpin at the C-terminus and excludes all other proteins with similar topology; similar domain fusions connect Dak and DegV, and genomic neighborhood organizations connect Dak and EIIA-man. Finally, both Dak and EIIA-man perform similar phosphotransfer reactions, suggesting a phosphotransferase activity for the DegV-like family of proteins, whose function other than lipid binding revealed in the crystal structure remains unknown. PDF
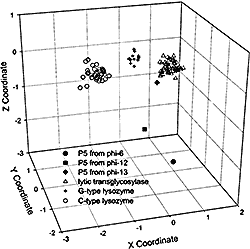
Peptidases are classical objects of enzymology and structural studies. However, a few protein families with experimentally characterized proteolytic activity, but unknown catalytic mechanism and three-dimensional structures, still exist. Using comparative sequence analysis, we deduce spatial structure for one of such families, namely, U40, which contains just one P5 protein from bacteriophage phi-6. We show that this singleton sequence possesses conserved sequence motifs characteristic of lysozymes and is a distant homolog of lytic transglycosylases that cleave bacterial peptidoglycan. The structure of the P5 protein is therefore predicted to adopt the lysozyme-like fold shared by T4, lambda, C-type, G-type lysozymes, and lytic transglycosylases. Since previous biochemical experiments with P5 of phi-6 have indicated that the purified enzyme possesses endopeptidase activity and not glycosidase activity, our results point to the possibility of a newly evolved molecular function and call for further experimental characterization of this unusual P5 protein. PDF
We also identified two new lysozyme-like protein families by using a combination of sequence similarity searches, domain architecture analysis, and structural predictions. First, the P5 protein from bacteriophage phi8, which belongs to COG3926 and Pfam family DUF847, is predicted to have a new lysozyme-like domain. This assignment is consistent with the lytic function of P5 proteins observed in several related double-stranded RNA bacteriophages. Domain architecture analysis reveals two lysozyme-associated transmembrane modules (LATM1 and LATM2) in a few COG3926/DUF847 members. LATM2 is also present in two proteins containing a peptidoglycan binding domain (PGB) and an N-terminal region that corresponds to COG5526 with uncharacterized function. Second, structure prediction and sequence analysis suggest that COG5526 represents another new lysozyme-like family. Our analysis offers fold and active-site assignments for COG3926/DUF847 and COG5526. The predicted enzymatic activity is consistent with an experimental study on the zliS gene product from Zymomonas mobilis, suggesting that bacterial COG3926/DUF847 members might be activators of macromolecular secretion. PDF
Understanding relationships between sequence, structure, and evolution is important for functional characterization of proteins. Here, we define a novel DOM-fold as a consensus structure of the domains in DmpA (L-aminopeptidase D-Ala-esterase/amidase), OAT (ornithine acetyltransferase), and MocoBD (molybdenum cofactor-binding domain), and discuss possible evolutionary scenarios of its origin. As shown by a comprehensive structure similarity search, DOM-fold distinguished by a two-layered beta/alpha architecture of a particular topology with unusual crossing loops is unique to those three protein families. DmpA and OAT are evolutionarily related as indicated by their sequence, structural, and functional similarities. Structural similarity between the DmpA/OAT superfamily and the MocoBD domains has not been reported before. Contrary to previous reports, we conclude that functional similarities between DmpA/OAT proteins and N-terminal nucleophile (Ntn) hydrolases are convergent and are unlikely to be inherited from a common ancestor. PDF
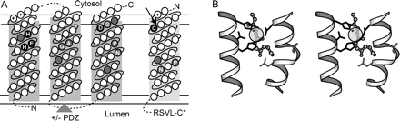
Site-2 proteases (S2Ps) form a large family of membrane-embedded metalloproteases that participate in cellular signaling pathways through sequential cleavage of membrane-tethered substrates. Using sequence similarity searches, we extend the S2P family to include remote homologs that help define a conserved structural core consisting of three predicted transmembrane helices with traditional metalloprotease functional motifs and a previously unrecognized motif (GxxxN/S/G). S2P relatives were identified in genomes from Bacteria, Archaea, and Eukaryota including protists, plants, fungi, and animals. The diverse S2P homologs divide into several groups that differ in various inserted domains and transmembrane helices. Mammalian S2P proteases belong to the major ubiquitous group and contain a PDZ domain. Sequence and structural analysis of the PDZ domain support its mediating the sequential cleavage of membrane-tethered substrates. Finally, conserved genomic neighborhoods of S2P homologs allow functional predictions for PDZ-containing transmembrane proteases in extra-cytoplasmic stress response and lipid metabolism. PDF
Restriction endonucleases and other nucleic acid cleaving enzymes form a large and extremely diverse superfamily that display little sequence similarity despite retaining a common core fold responsible for cleavage. The lack of significant sequence similarity between protein families makes homology inference a challenging task and hinders new family identification with traditional sequence-based approaches. Using the consensus fold recognition method Meta-BASIC that combines sequence profiles with predicted protein secondary structure, we identify nine new restriction endonuclease-like fold families among previously uncharacterized proteins and predict these proteins to cleave nucleic acid substrates. Application of transitive searches combined with gene neighborhood analysis allow us to confidently link these unknown families to a number of known restriction endonuclease-like structures and thus assign folds to the uncharacterized proteins. Finally, our method identifies a novel restriction endonuclease-like domain in the C-terminus of RecC that is not detected with structure-based searches of the existing PDB database. PDF
Eukaryotic protein trafficking pathways require specific transfer of cargo vesicles to different target organelles. A number of vesicle trafficking and membrane fusion components participate in this process, including various tethering factor complexes that interact with small GTPases prior to SNARE-mediated vesicle fusion. In Saccharomyces cerevisiae a protein complex of Mon1 and Ccz1 functions with the small GTPase Ypt7 to mediate vesicle trafficking to the vacuole. Mon1 belongs to DUF254 found in a diverse range of eukaryotic genomes, while Ccz1 includes a CHiPS domain that is also present in a known human protein trafficking disorder gene (HPS-4). We identify the CHiPS domain and a sequence region from another trafficking disorder gene (HPS-1) as homologs of an N-terminal domain from DUF254. This link establishes the evolutionary conservation of a protein complex (HPS-1/HPS-4) that functions similarly to Mon1/Ccz1 in vesicle trafficking to lysosome-related organelles of diverse eukaryotic species. Furthermore, the newly identified DUF254 domain is a distant homolog of the mu-adaptin longin domain found in clathrin adapter protein (AP) complexes of known structure that function to localize cargo protein to specific organelles. In support of this fold assignment, known longin domains such as the AP complex sigma-adaptin, the synaptobrevin N-terminal domains sec22 and Ykt6, and the srx domain of the signal recognition particle receptor also regulate vesicle trafficking pathways by mediating SNARE fusion, recognizing specialized compartments, and interacting with small GTPases that resemble Ypt7. PDF
Fic domains are found in a variety of species, including bacteria, a few archaea, and metazoan eukaryotes. The Vibrio parahaemolyticus type III secreted effector VopS contains a fic domain that covalently modifies Rho GTPase threonine with AMP to inhibit downstream signaling events in host cells. The VopS fic domain includes a conserved sequence motif (HPFx[D/E]GN[G/K]R) that contributes to AMPylation. We show that the AMPylation activity extends to a eukaryotic fic domain in Drosophila melanogaster CG9523, and use sequence and structure based computational methods to identify related domains in doc toxins and the type III effector AvrB. The conserved sequence motif that contributes to AMPylation unites fic with doc. Although AvrB lacks this motif, its structure reveals a similar topology to the fic and doc folds. AvrB binds to a peptide fragment of its host virulence target in a similar manner as fic binds peptide substrate. AvrB also orients a phosphate group from a bound ADP ligand near the peptide-binding site and in a similar position as a bound fic phosphate. The demonstrated eukaryotic fic domain AMPylation activity suggests that the VopS effector has exploited a novel host posttranslational modification. Fic domain-related structures give insight to the AMPylation active site and to the VopS fic domain interaction with its host GTPase target. These results suggest that fic, doc, and AvrB stem from a common ancestor that has evolved to AMPylate protein substrates. PDF
Most core components of the neurotransmitter release machinery have homologs in other types of intracellular membrane traffic, likely underlying a universal mechanism of intracellular membrane fusion. However, no clear similarity between Munc13 and protein families generally involved in membrane traffic has been reported, despite the essential nature of Munc13s for neurotransmitter release. This crucial function was ascribed to a minimal Munc13 region called the MUN domain, which likely participates in soluble N-ethylmaleimide sensitive factor attachment protein receptor complex (SNARE) assembly and is also found in Ca2+-dependent activator protein for secretion. We have now used comparative sequence and structural analyses to study the structure and evolutionary origin of the MUN domain. We found weak yet significant sequence similarities between the MUN domain and a set of protein subunits from several related vesicle tethering complexes, such as Sec6 from the exocyst complex and Vps53 from the Golgi-associated retrograde protein complex. Such an evolutionary relationship allows structure prediction of the MUN domain and suggests functional similarities between MUN domain-containing proteins and multisubunit tethering complexes such as exocyst, conserved oligomeric Golgi complex, Golgi-associated retrograde protein complex, and Dsl1p. These findings further unify the mechanism of neurotransmitter release with those of other types of intracellular membrane traffic and, in turn, support a role for tethering complexes in soluble N-ethylmaleimide sensitive factor attachment protein receptor complex assembly. Clearly, much research will be required to fully understand how MUN domains and tethering complexes function, but the connection established here leads to multiple predictions that can now be tested experimentally. For instance, it will be interesting to examine whether fragments of the MUN domain corresponding to the individual domains of tethering complexes of known structures can fold autonomously. Such fragments would help to figure out the sequences of the MUN domain that are involved in different types of interactions, including interactions with the SNAREs or putative intramolecular interactions with the Munc13-1 N-terminus that could provide a link to RIMs and Rab3. Moreover, it will also be interesting to investigate whether interactions mapped within the MUN domain are conserved in tethering complexes, or vice versa. These experiments will be crucial to further understand how conserved are the mechanisms of membrane docking and fusion at different membrane compartments. PDF
Tannerella forsythia is a bacterial pathogen involved in periodontal disease. A cysteine protease PrtH has been characterized in this bacterium as a virulence factor. PrtH has the activity of detaching adherent cells from substratum, and the level of PrtH is associated with periodontal attachment loss. No reports exist on the structure, active site, and catalytic mechanism of PrtH. Using comparative sequence and structural analyses, we have identified homologs of PrtH in a number of bacterial and archaeal species. PrtH was found to be remotely related to caspases and other proteases with a caspase-like fold, such as gingipains from another periodontal pathogen Porphyromonas gingivalis. The structures of caspases and gingipains have a Rossmann-fold like core characterized by a mainly parallel central β-sheet and surrounding α-helices on both sides. The structure of the cysteine protease domain from V. cholerae RTX toxin (peptidase family C80) exhibits a similar core as well as noticeable differences in peripheral regions as compared to the structures of caspases and gingipains. Conservation of the catalytic cysteine and histidine residues indicates that PrtH homologs also function as active proteolytic enzymes. The putative protease function of these proteins is also supported by genome context mining. Our results offer structural and mechanistic insights into PrtH and its homologs, and help classification of this protease family. PDF
CPDadh is a new peptidase family homologous to the cysteine protease domain in bacterial MARTX toxins. A cysteine protease domain (CPD) has been recently discovered in a group of multifunctional, autoprocessing RTX toxins (MARTX) and Clostridium difficile toxins A and B. These CPDs (referred to as CPDmartx) autocleave the toxins to release domains with toxic effects inside host cells. We report identification and computational analysis of CPDadh, a new cysteine peptidase family homologous to CPDmartx. CPDadh and CPDmartx share a Rossmann-like structural core and conserved catalytic residues. In bacteria, domains of the CPDadh family are present at the N-termini of a diverse group of putative cell-cell interaction proteins and at the C-termini of some RHS (recombination hot spot) proteins. In eukaryotes, catalytically inactive members of the CPDadh family are found in cell surface protein NELF (nasal embryonic LHRH factor) and some putative signaling proteins. The functions of CPDadh domains in different groups of bacterial and eukaryotic proteins remain to be understood. PDF
A Rho GTPase inactivation domain (RID) has been discovered in the multifunctional, autoprocessing RTX toxin RtxA from Vibrio cholerae. The RID domain causes actin depolymerization and rounding of host cells through inactivation of the small Rho GTPases Rho, Rac, and Cdc42. With only a few toxin proteins containing RID domains in the current sequence database, the structure and molecular mechanisms of this domain are unknown. Using comparative sequence and structural analyses, we report homology inference, fold recognition, and active site prediction for RID domains. Remote homologs of RID domains were identified in two other experimentally characterized bacterial virulence factors: IcsB of Shigella flexneri and BopA of Burkholderia pseudomallei, as well as in a group of uncharacterized bacterial membrane proteins. IcsB plays an important role in helping Shigella to evade the host autophagy defense system. RID domain homologs share a conserved diad of cysteine and histidine residues, and are predicted to adopt a circularly permuted papain-like thiol protease fold. RID domains from MARTX toxins and virulence factors IcsB and BopA thus could function as proteases or acyltransferases acting on host molecules. For example, one possible mechanism of the RID domain could be inactivation of GEFs by proteolytic cleavage. The substrate(s) of the RID domain remain to be experimentally discovered. The presence of a membrane-binding helical domain just N-terminal to the RID domain in MARTX toxins suggests that the RID substrate(s) might have a membrane localization. Our results provide structural and mechanistic insights into several important proteins functioning in bacterial pathogenesis. PDF
Candidatus Liberibacter asiaticus (Ca. L. asiaticus) is a Gram-negative bacterium and the pathogen of Citrus Greening disease (Huanglongbing, HLB). As a parasitic bacterium, Ca. L. asiaticus harbors ABC transporters that play important roles in exchanging chemical compounds between Ca. L. asiaticus and its host. We analyzed all the ABC transporter-related proteins in Ca. L. asiaticus. We identified 14 ABC transporter systems and predicted their structures and substrate specificities. In-depth sequence and structure analysis including multiple sequence alignment, phylogenetic tree reconstruction, and structure comparison further support their function predictions. Our study shows that this bacterium could use these ABC transporters to import metabolites (amino acids and phosphates) and enzyme cofactors (choline, thiamine, iron, manganese, and zinc), resist to organic solvent, heavy metal, and lipid-like drugs, maintain the composition of the outer membrane (OM), and secrete virulence factors. Although the features of most ABC systems could be deduced from the abundant experimental data on their orthologs, we reported several novel observations within ABC system proteins. Moreover, we identified seven nontransport ABC systems that are likely involved in virulence gene expression regulation, transposon excision regulation, and DNA repair. Our analysis reveals several candidates for further studies to understand and control the disease, including the type I virulence factor secretion system and its substrate that are likely related to Ca. L. asiaticus pathogenicity and the ABC transporter systems responsible for bacterial OM biosynthesis that are good drug targets. PDF
The heme-copper oxidase (HCO) superfamily includes HCOs in aerobic respiratory chains and nitric oxide reductases (NORs) in the denitrification pathway. The HCO/NOR catalytic subunit has a core structure consisting of 12 transmembrane helices (TMHs) arranged in three-fold rotational pseudosymmetry, with six conserved histidines for heme and metal binding. Using sensitive sequence similarity searches, we detected a number of novel HCO/NOR homologs and named them HCO Homology (HCOH) proteins. Several HCOH families possess only four TMHs that exhibit the most pronounced similarity to the last four TMHs (TMHs 9-12) of HCOs/NORs. Encoded by independent genes, four-TMH HCOH proteins represent a single evolutionary unit (EU) that relates to each of the three homologous EUs of HCOs/NORs comprising TMHs 1-4, TMHs 5-8, and TMHs 9-12. Single-EU HCOH proteins could form homotrimers or heterotrimers to maintain the general structure and ligand-binding sites defined by the HCO/NOR catalytic subunit fold. The remaining HCOH families, including NnrS, have twelve TMHs and three EUs. Most three-EU HCOH proteins possess two conserved histidines and could bind a single heme. Limited experimental studies and genomic context analysis suggest that many HCOH proteins could function in the denitrification pathway and in detoxification of reactive molecules such as nitric oxide. HCO/NOR catalytic subunits exhibit remarkable structural similarity to the homotrimers of MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism) proteins. Gene duplication, fusion, and fission likely play important roles in the evolution of HCOs/NORs and HCOH proteins. PDF
A large family of G protein-coupled receptors (GPCRs) involved in cell adhesion has a characteristic autoproteolysis motif of HLT/S known as the GPCR proteolysis site (GPS). Recent structural studies have elucidated the GPS to be part of a larger domain named GPCR autoproteolysis inducing (GAIN) domain. We demonstrated the remote homology relationships of GAIN domain to ZU5 domain and Nucleoporin98 (Nup98) C-terminal domain by structural and sequence analysis. Sequence homology searches were performed to extend ZU5-like domains to bacteria and archaea, as well as new eukaryotic families. Current diverse families of GAIN domain subdomain B, ZU5 and Nup98 C-terminal domain likely evolved from an ancient autoproteolytic domain with an HFS motif. The autoproteolytic site was kept intact in Nup98, PIDD and UNC5C-like, deteriorated in many ZU5 domains and changed in GAIN and FIIND. Deletion of the strand after the cleavage site was observed in ZO-1 and some Nup98 homologs. These findings link several autoproteolytic domains, extend our understanding of GAIN domain origination in adhesion GPCRs and provide insights into the evolution of an ancient autoproteolytic domain. PDF
CREST superfamily of transmembrane proteins. A number of membrane-spanning proteins possess enzymatic activity and catalyze important reactions involving proteins, lipids or other substrates located within or near lipid bilayers. Alkaline ceramidases are seven-transmembrane proteins that hydrolyze the amide bond in ceramide to form sphingosine. Recently, a group of putative transmembrane receptors called progestin and adipoQ receptors (PAQRs) were found to be distantly related to alkaline ceramidases, raising the possibility that they may also function as membrane enzymes. Using sensitive similarity search methods, we identified statistically significant sequence similarities among several transmembrane protein families including alkaline ceramidases and PAQRs. They were unified into a large and diverse superfamily of putative membrane-bound hydrolases called CREST (alkaline ceramidase, PAQR receptor, Per1, SID-1 and TMEM8). The CREST superfamily embraces a plethora of cellular functions and biochemical activities, including putative lipid-modifying enzymes such as ceramidases and the Per1 family of putative phospholipases involved in lipid remodeling of GPI-anchored proteins, putative hormone receptors, bacterial hemolysins, the TMEM8 family of putative tumor suppressors, and the SID-1 family of putative double-stranded RNA transporters involved in RNA interference. Extensive similarity searches and clustering analysis also revealed several groups of proteins with unknown function in the CREST superfamily. Members of the CREST superfamily share seven predicted core transmembrane segments with several conserved sequence motifs. Universal conservation of a set of histidine and aspartate residues across all groups in the CREST superfamily, coupled with independent discoveries of hydrolase activities in alkaline ceramidases and the Per1 family as well as results from previous mutational studies of Per1, suggests that the majority of CREST members are metal-dependent hydrolases. PDF
The Shisa family of single-transmembrane proteins is characterized by an N-terminal cysteine-rich domain and a proline-rich C-terminal region. Its founding member, Xenopus Shisa, promotes head development by antagonizing Wnt and FGF signaling. Recently, a mouse brain-specific Shisa protein CKAMP44 (Shisa9) was shown to play an important role in AMPA receptor desensitization. We used sequence similarity searches against protein, genome and EST databases to study the evolutionary origin and phylogenetic distribution of Shisa homologs. In addition to nine Shisa subfamilies in vertebrates, we detected distantly related Shisa homologs that possess an N-terminal domain with six conserved cysteines. These Shisa-like proteins include FAM159 and KIAA1644 mainly from vertebrates, and members from various bilaterian invertebrates and Porifera, suggesting their presence in the last common ancestor of Metazoa. Shisa-like genes have undergone large expansions in Branchiostoma floridae and Saccoglossus kowalevskii, and appear to have been lost in certain insects. Pattern-based searches against eukaryotic proteomes also uncovered several other families of predicted single-transmembrane proteins with a similar cysteine-rich domain. We refer to these proteins (Shisa/Shisa-like, WBP1/VOPP1, CX, DUF2650, TMEM92, and CYYR1) as STMC6 proteins (single-transmembrane proteins with conserved 6 cysteines). STMC6 genes are widespread in Metazoa, with the human genome containing 17 members. Frequent occurrences of PY motifs in STMC6 proteins suggest that most of them could interact with WW-domain-containing proteins, such as the NEDD4 family E3 ubiquitin ligases, and could play critical roles in protein degradation and sorting. STMC6 proteins are likely transmembrane adaptors that regulate membrane proteins such as cell surface receptors. PDF
Cysteine-rich domains related to Frizzled receptors and Hedgehog-interacting proteins. Frizzled and Smoothened are homologous seven-transmembrane proteins functioning in the Wnt and Hedgehog signaling pathways, respectively. They harbor an extracellular cysteine-rich domain (FZ-CRD), a mobile evolutionary unit that has been found in a number of other metazoan proteins and Frizzled-like proteins in Dictyostelium. Domains distantly related to FZ-CRDs, in Hedgehog-interacting proteins (HHIPs), folate receptors and riboflavin-binding proteins (FRBPs), and Niemann-Pick Type C1 proteins (NPC1s), referred to as HFN-CRDs, exhibit similar structures and disulfide connectivity patterns compared with FZ-CRDs. We used computational analyses to expand the homologous set of FZ-CRDs and HFN-CRDs, providing a better understanding of their evolution and classification. First, FZ-CRD-containing proteins with various domain compositions were identified in several major eukaryotic lineages including plants and Chromalveolata, revealing a wider phylogenetic distribution of FZ-CRDs than previously recognized. Second, two new and distinct groups of highly divergent FZ-CRDs were found by sensitive similarity searches. One of them is present in the calcium channel component Mid1 in fungi and the uncharacterized FAM155 proteins in metazoans. Members of the other new FZ-CRD group occur in the metazoan-specific RECK (reversion-inducing-cysteine-rich protein with Kazal motifs) proteins that are putative tumor suppressors acting as inhibitors of matrix metalloproteases. Finally, sequence and three-dimensional structural comparisons helped us uncover a divergent HFN-CRD in glypicans, which are important morphogen-binding heparan sulfate proteoglycans. Such a finding reinforces the evolutionary ties between the Wnt and Hedgehog signaling pathways and underscores the importance of gene duplications in creating essential signaling components in metazoan evolution. PDF
KLF18 is a new gene/pseudogene family of Krüppel-like transcription factors. Krüppel-like factors (KLF) and specificity proteins (SP) constitute a family of zinc-finger-containing transcription factors that play important roles in a wide range of processes including differentiation and development of various tissues. The human genome possesses 17 KLF genes (KLF1-KLF17) and nine SP genes (SP1-SP9) with diverse functions. We used sequence similarity searches and gene synteny analysis to identify a new putative KLF gene/pseudogene named KLF18 that is present in most of the placental mammals with sequenced genomes. KLF18 is a chromosomal neighbor of the KLF17 gene and is likely a product of its duplication. Phylogenetic analyses revealed that mammalian predicted KLF18 proteins and KLF17 proteins experienced elevated rates of evolution and are grouped with KLF1/KLF2/KLF4 and non-mammalian KLF17. Predicted KLF18 proteins maintain conserved features in the zinc fingers of the SP/KLF family, while possessing repeats of a unique sequence motif in their N-terminal regions. No expression data have been reported for KLF18, suggesting that it either has highly restricted expression patterns and specialized functions, or could have become a pseudogene in extant placental mammals. Besides KLF18 genes/pseudogenes, we identified several KLF18-like genes such as Zfp352, Zfp352-like, and Zfp353 in the genomes of mouse and rat. These KLF18-like genes do not possess introns inside their coding regions, and gene expression data indicate that some of them may function in early embryonic development. They represent further expansions of KLF members in the murine lineage, most likely resulted from several events of retrotransposition and local gene duplication starting from an ancient spliced mRNA of KLF18. PDF
Specificity proteins (SPs) and Krüppel-like factors (KLFs) are C2H2-type Zinc finger transcription factors that play essential roles in differentiation, development, proliferation and cell death. SP/KLF proteins, similarly to Wilms tumor protein 1 (WT1), Early Growth Response (EGR), Huckebein, and Klumpfuss, prefer to bind GC-rich sequences such as GC-box and CACCC-box (GT-box). We searched various genomes and transcriptomes of metazoans and single-cell holozoans for members of these families and identified seven groups of KLFs (KLFA-G) and three groups of SPs (SPA-C) in the three lineages of Bilateria (Deuterostomia, Ecdysozoa, and Lophotrochozoa). The last ancestor of jawed vertebrates was inferred to have at least 18 KLFs. Multiple KLF members were found in basal metazoans (Ctenophora, Porifera, Placozoa, and Cnidaria). While SP, EGR and Klumpfuss were only detected in metazoans, KLF, WT1, and Huckebein are present in nonmetazoan holozoans. PDF
Type II CAAX protease families. Intramembrane proteases are responsible for a number of regulated proteolysis events occurring within or near the plasma and intracellular membranes. Members of one large and diverse family of putative intramembrane metalloproteases are widely distributed in all domains of life, including the type II CAAX prenyl proteases and their prokaryotic homologs with putative bacteriocin-related functions. We used sensitive sequence similarity searches to expand this large CPBP (CAAX proteases and bacteriocin-processing enzymes) family to include more than 5800 members and infer its homologous relationships to several other protein families, including the PrsW proteases, the DUF2324 (DUF, domain of unknown function) family and the gamma-secretase subunit APH-1 proteins. They share four predicted core transmembrane segments and possess similar yet distinct sets of sequence motifs. Remote similarity between APH-1 and membrane proteases sheds light on APH-1's evolutionary origin and raises the possibility that APH-1 may possess proteolytic activity in the current or ancestral form of gamma-secretase. PDF
HAP2, the eukaryotic ancient gamete fusogen, is a remote homolog of viral class II fusion proteins. Sexual reproduction, essential and nearly universal in eukaryotes, involves the fusion of male and female haploid gametes into a diploid cell. The sperm-restricted single-pass transmembrane protein HAP2-GCS1, a single-pass transmembrane protein present in sperm, has been proposed to function in membrane fusion. Its presence in the major eukaryotic taxa-animals, plants, and protists (including important human pathogens like Plasmodium)-suggests that many eukaryotic organisms share a common gamete fusion mechanism. We provided bioinformatic support of homology between HAP2 and class II viral membrane fusion proteins, and in collaboration with experimental researchers unveiled the structures of HAP2 of the unicellular alga Chlamydomonas reinhardtii. Targeting the segment corresponding to the fusion loop by mutagenesis or by antibodies blocks gamete fusion, demonstrating that HAP2 is the gamete fusogen with a mechanism of action akin to viral fusion. These results suggest a way to block Plasmodium transmission and highlight the impact of virus-cell genetic exchanges on the evolution of eukaryotic life. PDF
Complexins are synaptic SNARE complex-binding proteins that cooperate with synaptotagmins in activating Ca2+-stimulated, synaptotagmin?dependent synaptic vesicle exocytosis and in clamping spontaneous, synaptotagmin?independent synaptic vesicle exocytosis. Through bioinformatic studies, we show that complexin sequences are conserved in some non?metazoan unicellular organisms and in all metazoans, suggesting that complexins are a universal feature of metazoans that predate metazoan evolution. Experimental studies (performed in collaborator Dr. Thomas Südhof's lab) suggest that complexin from Nematostella vectensis, a cnidarian sea anemone far separated from mammals in metazoan evolution, functionally replaces mouse complexins in activating Ca2+-triggered exocytosis, but is unable to clamp spontaneous exocytosis. Thus, the activating function of complexins is likely conserved throughout metazoan evolution. PDF
SEA (sea urchin sperm protein, enterokinase, agrin) domains are found in a number of cell surface and secreted proteins, and many of them possess autoproteolysis activity. The presence of a SEA domain adjacent to the transmembrane segment appears to be a recurring theme in quite a number of type I transmembrane proteins on the cell surface, such as MUC1, dystroglycan, IA?2, and Notch receptors. By comparative sequence and structural analyses, we identified dystroglycan?like proteins with SEA domains in Capsaspora owczarzaki of the Filasterea group, one of the closest single?cell relatives of metazoans. We also detected novel and divergent SEA domains in a variety of cell surface proteins such as EpCAM, ?/??sarcoglycan, PTPRR, collectrin/Tmem27, amnionless, CD34, KIAA0319, fibrocystin?like protein, and a number of cadherins. While these proteins are mostly from metazoans or their single cell relatives such as choanoflagellates and Filasterea, fibrocystin?like proteins with SEA domains were found in several other eukaryotic lineages including green algae, Alveolata, Euglenozoa, and Haptophyta, suggesting an ancient evolutionary origin. In addition, the intracellular protein Nucleoporin 54 (Nup54) acquired a divergent SEA domain in choanoflagellates and metazoans.PDF
The Vibrio parahaemolyticus virulence type III secretion system (T3SS) is activated for secretion of bacterial toxins when the pathogen reaches the host gut. We assigned the Vp VtrC gene product as a distant member of the calycin superfamily, whose members bind lipid molecules at the center of their distinctive 8-stranded β-barrel meander fold. This detected homology suggested that VtrC could also bind a lipid molecule, such as bile found in the host gut. Indeed, an unusual structure of an obligate VtrC/VtrA heterodimer revealed the VtrC as a calycin that binds bile salts in the center of the barrel, thus activating the T3SS system while in the host gut.PDF
Studies of Citrus greening up
Candidatus Liberibacter asiaticus (Ca. L. asiaticus) is a parasitic gram-negative bacterium that is closely associated with Huanglongbing (HLB), a worldwide citrus disease. Given the difficulty in culturing the bacterium and thus in its experimental characterization, computational analyses of the whole Ca. L. asiaticus proteome can provide much needed insights into the mechanisms of the disease and guide the development of treatment strategies. We applied state-of-the-art sequence analysis tools to every Ca. L. asiaticus protein. Our results are available as a public website at http://prodata.swmed.edu/liberibacter_asiaticus/ In particular, we manually curated the results to predict the subcellular localization, spatial structure and function of all Ca. L. asiaticus proteins (http://prodata.swmed.edu/liberibacter_asiaticus/curated/). This extensive information should facilitate the study of Ca. L. asiaticus proteome function and its relationship to disease. Pilot studies based on the information from our website have revealed several potential virulence factors. PDF
Towards prediction of phenotype from genotype up
Predicting phenotype from genotype represents the epitome of biological questions. Comparative genomics of appropriate model organisms holds the promise of making it possible. To tackle this problem, we are developing methods and applying them to sequence, annotate, and analyze complete Eukaryotic genomes.
The fungal genus Stachybotrys produces several diverse toxins that affect human health. Its strains comprise two mutually-exclusive toxin chemotypes, one producing satratoxins, which are a subclass of trichothecenes, and the other producing the less-toxic atranones. To determine the genetic basis for chemotype-specific differences in toxin production, the genomes of four Stachybotrys strains were sequenced and assembled de novo. Two of these strains produce atranones and two produce satratoxins. Comparative analysis of these four 35-Mbp genomes revealed several chemotype-specific gene clusters that are predicted to make secondary metabolites. The largest, which was named the core atranone cluster, encodes 14 proteins that may suffice to produce all observed atranone compounds via reactions that include an unusual Baeyer-Villiger oxidation. Satratoxins are suggested to be made by products of multiple gene clusters that encode 21 proteins in all, including polyketide synthases, acetyltransferases, and other enzymes expected to modify the trichothecene skeleton. One such satratoxin chemotype-specific cluster is adjacent to the core trichothecene cluster, which has diverged from those of other trichothecene producers to contain a unique polyketide synthase. The results suggest that chemotype-specific gene clusters are likely the genetic basis for the mutually-exclusive toxin chemotypes of Stachybotrys. A unified biochemical model for Stachybotrys toxin production is presented. Overall, the four genomes described here will be useful for ongoing studies of this mold's diverse toxicity mechanisms. PDF
The bowhead whale (Balaena mysticetus) is estimated to live over 200 years and is possibly the longest-living mammal. These animals should possess protective molecular adaptations relevant to age-related diseases, particularly cancer. In collaboration with the de Magalhães group, we sequenced the bowhead whale genome and two transcriptomes from different populations. Our analysis identifies genes under positive selection and bowhead-specific mutations in genes linked to cancer and aging. In addition, we identify gene gain and loss involving genes associated with DNA repair, cell-cycle regulation, cancer, and aging. Our results expand our understanding of the evolution of mammalian longevity and suggest possible players involved in adaptive genetic changes conferring cancer resistance. We also found potentially relevant changes in genes related to additional processes, including thermoregulation, sensory perception, dietary adaptations, and immune response. Our data are made available online (http://www.bowhead-whale.org) to facilitate research in this long-lived species. PDF
The high heterozygosity of many Eukaryotes currently prohibits assembling their genomes. We obtained the 376 Mb genome sequence of Eastern Tiger Swallowtail Papilio glaucus (Pgl), the first sequenced genome from the Papilionidae family. We obtained the genome from a wild-caught specimen using a cost-effective strategy that overcomes the high (2%) heterozygosity problem. Comparative analyses suggest the molecular bases of various phenotypic traits, including terpene production in the Papilionidae-specific organ, osmeterium. Comparison of Pgl and Papilio canadensis transcriptomes reveals mutation hotspots (4% genes) associated with their divergence: four key circadian clock proteins are enriched in interspecies mutations and likely responsible for the difference in pupal diapause. Finally, the Pgl genome confirms Papilio appalachiensis as a hybrid of Pgl and Pca, but suggests it inherited 3/4 of its genes from Pca. PDF
Macroevolution of butterflies up
For centuries, biologists have used phenotypes to infer evolution. For decades, a handful of gene markers have given us a glimpse of the genotype to combine with phenotypic traits. Today, we can sequence entire genomes from hundreds of species and gain yet closer scrutiny. To illustrate the power of genomics, we have chosen skipper butterflies (Hesperiidae). The genomes of 250 representative species of skippers reveal rampant inconsistencies between their current classification and a genome-based phylogeny. We use a dated genomic tree to define tribes (six new) and subtribes (six new), to overhaul genera (nine new) and subgenera (three new), and to display convergence in wing patterns that fooled researchers for decades. We find that many skippers with similar appearance are only distantly related. This likely mimetic convergence is diversified, resulting in five distinct parallel wing patterns. Each of the five patterns occurs within at least two genera as well as in more distant relatives diverged more than 20 Mya. Conversely, we see that several skippers with distinct morphology are close relatives. Our conclusions are strongly supported by different genomic regions and are consistent with some morphological traits. Our work is a forerunner to genomic biology shaping biodiversity research. PDF
Biologists marvel at the powers of adaptive convergence, when distantly related animals look alike. While mimetic wing patterns of butterflies have fooled predators for millennia, entomologists inferred that mimics were distant relatives despite similar appearance. However, the obverse question has not been frequently asked. Who are the close relatives of mimetic butterflies and what are their features? As opposed to close convergence, divergence from a non-mimetic relative would also be extreme. When closely related animals look unalike, it is challenging to pair them. Genomic analysis promises to elucidate evolutionary relationships and shed light on molecular mechanisms of divergence. We chose the firetip skipper butterfly as a model due to its phenotypic diversity and abundance of mimicry. We sequenced and analysed whole genomes of nearly 120 representative species. Genomes partitioned this subfamily Pyrrhopyginae into five tribes (1 new), 23 genera and, additionally, 22 subgenera (10 new). The largest tribe Pyrrhopygini is divided into four subtribes (three new). Surprisingly, we found five cases where a uniquely patterned butterfly was formerly placed in a genus of its own and separately from its close relatives. In several cases, extreme and rapid phenotypic divergence involved not only wing patterns but also the structure of the male genitalia. The visually striking wing pattern difference between close relatives frequently involves disappearance or suffusion of spots and colour exchange between orange and blue. These differences (in particular, a transition between unspotted black and striped wings) happen recurrently on a short evolutionary time scale, and are therefore probably achieved by a small number of mutations. PDF
Since its accidental introduction to Massachusetts in the late 1800s, the European gypsy moth (EGM; Lymantria dispar dispar) has become a major defoliator in North American forests. However, in part because females are flightless, the spread of the EGM across the United States and Canada has been relatively slow over the past 150 years. In contrast, females of the Asian gypsy moth (AGM; Lymantria dispar asiatica) subspecies have fully developed wings and can fly, thereby posing a serious economic threat if populations are established in North America. To explore the genetic determinants of these phenotypic differences, in collaboration with the Gammon lab, we sequenced and annotated a draft genome of gypsy moth (L. dispar) and used it to identify genetic variation between EGM and AGM populations. The 865-Mb gypsy moth genome is the largest Lepidoptera genome sequenced to date and encodes ~13,300 proteins. Gene ontology analyses of EGM and AGM samples revealed divergence between these populations in genes enriched for several gene ontology categories related to muscle adaptation, chemosensory communication, detoxification of food plant foliage, and immunity. These genetic differences likely contribute to variations in flight ability, chemical sensing, and pathogen interactions among EGM and AGM populations. Finally, we use our new genomic and transcriptomic tools to provide insights into genome-wide gene-expression changes of the gypsy moth after viral infection. Characterizing the immunological response of gypsy moths to virus infection may aid in the improvement of virus-based bioinsecticides currently used to control larval populations. PDF
Studies of life rely on classifying organisms into species. Contrary to a frequent belief, simple and quantitative standards for species delineation are lacking, and debates about species delimitation create obstacles for conservation biology, agriculture, legislation, and education. To tackle this key biological question, we have chosen butterflies as model organisms. We sequenced and analyzed transcriptomes of 186 butterfly specimens representing pairs of close but clearly distinct species, conspecific populations, and taxa that are debated among experts. We find that species are robustly separated from conspecific populations by the combination of two measures computed on Z-linked genes: fixation index that detects hiatus between species, and the extent of gene flow that quantifies reproductive isolation. These criteria suggest that all 9 butterfly pairs that caused experts' disagreement are distinct species, not populations or subspecies. When applied to Homo, our criteria agree that all modern humans are the same species distinct from Neanderthals, suggesting relevance of this study beyond butterflies. Furthermore, we found that divergence and positive selection in proteins involved in interaction with DNA (including proteins encoded by trans-regulatory elements), circadian clock, pheromone sensing, development, and immune response recurrently correlate with speciation. A significant fraction of these divergent proteins is encoded by the Z chromosome, which appears to be more resistant to introgression than autosomes. Taken together, we find possible common speciation mechanisms in butterflies, present additional support for an important role of the Z chromosome in speciation of butterflies, and suggest quantitative criteria for butterfly species delimitation using genomic data, which is vital for the exploration of biodiversity. PDF
Never before have we had the luxury of choosing a continent, picking a large phylogenetic group of animals, and obtaining genomic data for its every species. We sequenced all 845 species of butterflies recorded from North America north of Mexico. Our comprehensive approach reveals the pattern of diversification and adaptation occurring in this phylogenetic lineage as it has spread over the continent, which cannot be seen on a sample of selected species. We observe bursts of diversification that generated taxonomic ranks: subfamily, tribe, subtribe, genus, and species. The older burst around 70 Mya resulted in the butterfly subfamilies, with the major evolutionary inventions being unique phenotypic traits shaped by high positive selection and gene duplications. The recent burst around 5 Mya is caused by explosive radiation in diverse butterfly groups associated with diversification in transcription and mRNA regulation, morphogenesis, and mate selection. Rapid radiation correlates with more frequent introgression of speciation-promoting and beneficial genes among radiating species. Radiation and extinction patterns over the last 100 million years suggest the following general model of animal evolution. A population spreads over the land, adapts to various conditions through mutations, and diversifies into several species. Occasional hybridization between these species results in accumulation of beneficial alleles in one, which eventually survives, while others become extinct. Not only butterflies, but also the hominids may have followed this path. PDF
Traditionally, animal classification was carried out based on morphology. Higher-level taxa, i.e., those above species level, such as genus, tribe or subfamily are usually defined as prominent clades in animal phylogeny. Genomic sequences are expected to give us the ultimate knowledge of phylogenetic relationships and thus genome-level phylogeny should be most accurate to use for higher classification. Whole genome shotgun sequences or butterflies are straightforward to convert to phylogenetic trees constructed from protein-coding regions. Inspection of these trees frequently reveals their incongruence with the currently used classification of butterflies. We correct these inconsistencies, and this work frequently results in the discovery of new taxa. For instance, we delineated three new subfamilies of skipper butterflies (Hesperiidae), reclassified the Emesidini tribe of metalmark butterflies, improved the classification of butterfly species found in the USA and Canada, and described 50 new genera of Hesperiidae. To learn more about thes work, consult the PDFs of papers here: PDF1 PDF2 PDF3 PDF4
Sequence and profile similarity search up
We developed a novel method for the comparison of multiple protein alignments with assessment of statistical significance (COMPASS). The method derives numerical profiles from alignments, constructs optimal local profile-profile alignments and analytically estimates E-values for the detected similarities. The scoring system and E-value calculation are based on a generalization of the PSI-BLAST approach to profile-sequence comparison, which is adapted for the profile-profile case. Tested along with existing methods for profile-sequence (PSI-BLAST) and profile-profile (prof_sim) comparison, COMPASS shows increased abilities for sensitive and selective detection of remote sequence similarities, as well as improved quality of local alignments. The method allows prediction of relationships between protein families in the PFAM database beyond the range of conventional methods. Two predicted relations with high significance are similarities between various Rossmann-type folds and between various helix-turn-helix-containing families. The potential value of COMPASS for structure/function predictions is illustrated by the detection of an intricate homology between the DNA-binding domain of the CTF/NFI family and the MH1 domain of the Smad family. COMPASS server is available here. Recentry, we modified the server that now features three major developments: (i) improved statistical accuracy; (ii) increased speed from parallel implementation; and (iii) new functional features facilitating structure prediction. These features include visualization tools that allow the user to quickly and effectively analyze specific local structural region predictions suggested by COMPASS alignments. For the top significant hits in databases of proteins with known 3D structure, an additional panel is displayed in the alignment section. First, a Jmol panel is used to interactively display the Cα trace of the structural fragment covered by the COMPASS alignment. Second, the user can analyze the structure in more detail, either by downloading the fragment as an all-atom PDB file, or by clicking on the ‘PyMOL’ link, which generates and launches a PyMOL script to show, in a separate window, the full structure of the potential homolog, with the aligned region highlighted. PDF1 PDF2 PDF3 PDF4
Profile-profile comparison is aimed at detecting similarities in amino acid preferences at sequence positions in two distant families. In addition to the residue substitution preferences (‘vertical’ signals), MSA can reveal patterns of interdependence between amino acid content at different positions (‘horizontal’ signals). These patterns, dictated by structure and function, are often preserved better than the sequence and thus can help detecting protein similarity where individual sequence positions diverged beyond recognition. Currently, such ‘horizontal’ information is used by only a few methods, mainly in the form of secondary structure (SS) prediction. Building on the COMPASS platform we engineered ProCAIn, a program for MSA comparison based on the combination of ‘vertical’ MSA context (substitution constraints at individual sequence positions) and ‘horizontal’ context (patterns of residue content at multiple positions). Based on a simple and tractable profile methodology and primitive measures for the similarity of horizontal MSA patterns, the method achieves the quality of homology detection comparable to a more complex advanced method employing hidden Markov models (HMMs) and secondary structure (SS) prediction. Adding SS information further improves PROCAIN performance beyond the capabilities of current state-of-the-art tools. In addition to using 'horizontal' information, we applied a non-traditional way to estimate statistical significance of the results. Usually, a distribution of database hit scores for a given query is used to estimate E-value. However it is challenging to remove homologs from the hit list, because some homologs may have very low scores. Therefore it is not easy to obtain a distribution of scores for non-homologous hits to a query. However, if the database consists of proteins with known structures, and for structure prediction applications this is the case, we can confidently find all proteins in the database with high structural similarity to a hit (=subject). Thus for each query-subject pair, a distribution of random scores (scores to structurally similar proteins are removed from the sample) to a subject, rather than a query, can be used to obtain the estimate of statistical significance. Such statistics built on subjects rather than on the query result in better ranking of hits. Moreover, being combined with the statistics computed on the query compared to a calibration database of mostly non-homologous domains, subject-based statistics are further improved. The potential value of the ProCAIn method for structure-function predictions is illustrated by the detection of subtle homology between evolutionary distant yet structurally similar protein domains. ProCAIn, relevant databases and tools can be downloaded from here. The web server can be accessed here. PDF1 PDF2 PDF3
Profile-based similarity search is an essential step in structure-function studies of proteins. However, inclusion of non-homologous sequence segments into a profile causes its corruption and results in false positives. Profile corruption is common in multidomain proteins. One major source of profile corruption is from extension of alignments over non-homologous sequence regions. For instance, for two 2-domain proteins, AB and A’C, PSI-BLAST may extend a correct alignment of the homologous domains A and A’ to include sequences from the non-homologous domains B and C. Despite significant effort devoted to this multi-domain problem, no satisfactory solution exists. Currently, the best approach is to start PSI-BLAST with precisely defined query sequence bounds However, we found that even a single, well-defined domain does not guarantee a corruption-free profile. Domains hosting insertions, which represent close to 5% of domains in the SCOP 1.75 database, may generate a corrupted PSI-BLAST profile due to incorrect alignment extension around the insertion position. We developed a procedure (HangOut) that, for a single domain with specified insertion position, cleans erroneously extended PSI-BLAST alignments to generate better profiles. PSI-BLAST alignment can be divided into two segments: (1) correctly aligned and (2) incorrectly aligned or extended. These incorrectly aligned “overhangs” are detected and removed by the HangOut program to clean the profile and prepare it for consequent remote homology searches with various tools, such as PSI-BLAST and HHsearch. When tested on all 302 SCOP domains harboring inserted domain. PSI-BLAST profiles show corruption in 30% cases (91 domains). All but one HangOut profiles are free from corruption. The single exception is due to distant homology, since both the host and inserted domain represent similar doubly wound Rossmann folds. Although our current HangOut method does not offer a comprehensive solution to the multidomain problem, it addresses a special case of domains with insertions that represent the major source of profile corruption when PSI-BLAST is initiated with single, discontinuous domain queries.
Clustering of proteins up
Biological objects tend to cluster into discrete groups. Objects within a group typically possess similar properties. It is important to have fast and efficient tools for grouping objects that result in biologically meaningful clusters. Protein sequences reflect biological diversity and offer an extraordinary variety of objects for polishing clustering strategies. Grouping of sequences should reflect their evolutionary history and their functional properties. Visualization of relationships between sequences is of no less importance. Tree-building methods are typically used for such visualization. An alternative concept to visualization is a multidimensional sequence space. In this space, proteins are defined as points and distances between the points reflect the relationships between the proteins. Such a space can also be a basis for model-based clustering strategies that typically produce results correlating better with biological properties of proteins. We developed an approach to classification of biological objects that combines evolutionary measures of their similarity with a model-based clustering procedure. We apply the methodology to amino acid sequences. On the first step, given a multiple sequence alignment, we estimate evolutionary distances between proteins measured in expected numbers of amino acid substitutions per site. These distances are additive and are suitable for evolutionary tree reconstruction. On the second step, we find the best fit approximation of the evolutionary distances by Euclidian distances and thus represent each protein by a point in a multidimensional space. The Euclidian space may be projected in two or three dimensions and the projections can be used to visualize relationships between proteins. On the third step, we find a non-parametric estimate of the probability density of the points and cluster the points that belong to the same local maximum of this density in a group. The number of groups is controlled by a sigma-parameter that determines the shape of the density estimate and the number of maxima in it. The grouping procedure outperforms commonly used methods such as UPGMA and single linkage clustering. See http://prodata.swmed.edu/download/pub/EESG PDF
Inference of remote homology between proteins is very challenging and remains a prerogative of an expert. Thus a significant drawback to the use of evolutionary-based protein structure classifications is the difficulty in assigning new proteins to unique positions in the classification scheme with automatic methods. To address this issue, we have developed an algorithm to map protein domains to an existing structural classification scheme and have applied it to the SCOP database. The general strategy employed by this algorithm is to combine the results of several existing sequence and structure comparison tools applied to a query protein of known structure in order to find the homologs already classified in SCOP database and thus determine classification assignments. The algorithm is able to map domains within newly solved structures to the appropriate SCOP superfamily level with approximately 95% accuracy. Examples of correctly mapped remote homologs are discussed. The algorithm is also capable of identifying potential evolutionary relationships not specified in the SCOP database, thus helping to make it better. The strategy of the mapping algorithm is not limited to SCOP and can be applied to any other evolutionary-based classification scheme as well. SCOPmap is available for download. The SCOPmap program is useful for assigning domains in newly solved structures to appropriate superfamilies and for identifying evolutionary links between different superfamilies. PDF
Secondary structure delineation up
The majority of residues in protein structures are involved in the formation of alpha-helices and beta-strands. These distinctive secondary structure patterns can be used to represent a protein for visual inspection and in vector-based protein structure comparison. Success of such structural comparison methods depends crucially on the accurate identification and delineation of secondary structure elements. We have developed a method PALSSE (Predictive Assignment of Linear Secondary Structure Elements) that delineates secondary structure elements (SSEs) from protein Cα coordinates and specifically addresses the requirements of vector-based protein similarity searches. Our program identifies two types of secondary structures: helix and β-strand, typically those that can be well approximated by vectors. In contrast to traditional secondary structure algorithms, which identify a secondary structure state for every residue in a protein chain, our program attributes residues to linear SSEs. Consecutive elements may overlap, thus allowing residues located at the overlapping region to have more than one secondary structure type. PALSSE is predictive in nature and can assign about 80% of the protein chain to SSEs as compared to 53% by DSSP and 57% by P-SEA. Such a generous assignment ensures almost every residue is part of an element and is used in structural comparisons. Our results are in agreement with human judgment and DSSP. The method is robust to coordinate errors and can be used to define SSEs even in poorly refined and low-resolution structures. The program and results are available at http://prodata.swmed.edu/palsse. PDF
3D structure pattern search up
Many evolutionarily distant, but functionally meaningful links between proteins come to light through comparison of spatial structures. Most programs that assess structural similarity compare two proteins to each other and find regions in common between them. Structural classification experts look for a particular structural motif instead. Programs base similarity scores on superposition or closeness of either Cartesian coordinates or inter-residue contacts. Experts pay more attention to the general orientation of the main chain and mutual spatial arrangement of secondary structural elements. There is a need for a computational tool to find proteins with the same secondary structures, topological connections and spatial architecture, regardless of subtle differences in 3D coordinates. We developed ProSMoS – a Protein Structure Motif Search program that emulates an expert. Starting from a spatial structure, the program uses previously delineated secondary structural elements. A meta-matrix of interactions between the elements (parallel or antiparallel) minding handedness of connections (left or right) and other features (e.g. element lengths and hydrogen bonds) is constructed prior to or during the searches. All structures are reduced to such meta-matrices that contain just enough informatiPDZon to define a protein fold, but this definition remains very general and deviations in 3D coordinates are tolerated. User supplies a meta-matrix for a structural motif of interest, and ProSMoS finds all proteins in the protein data bank (PDB) that match the meta-matrix. ProSMoS performance is compared to other programs and is illustrated on a beta-Grasp motif. A brief analysis of all beta-Grasp-containing proteins is presented. Program availability: ProSMoS is freely available for non-commercial use from http://prodata.swmed.edu/download/pub/ProSMoS. PDF
Multiple sequence alignment up
Although multiple sequence alignments (MSAs) are essential for a wide range of applications from structure modeling to prediction of functional sites, construction of accurate MSAs for distantly related proteins remains a largely unsolved problem. Over the last few years we paid significant attention to the problem and developed four MSA programs.
PCMA (profile consistency multiple sequence alignment) is a progressive multiple sequence alignment program that combines two different alignment strategies. Highly similar sequences are aligned in a fast way as in ClustalW, forming pre-aligned groups. The T-Coffee strategy is applied to align the relatively divergent groups based on profile-profile comparison and consistency. The scoring function for local alignments of pre-aligned groups is based on a novel profile-profile comparison method that is a generalization of the PSI-BLAST approach to profile-sequence comparison. PCMA balances speed and accuracy in a flexible way and is suitable for aligning large numbers of sequences. AVAILABILITY: PCMA is freely available for non-commercial use. Pre-compiled versions for several platforms can be downloaded from http://prodata.swmed.edu/download/pub/PCMA and a web-server is set up here. PDF
We have developed MUMMALS, a program to construct multiple protein sequence alignment using probabilistic consistency. MUMMALS improves alignment quality by using pairwise alignment hidden Markov models (HMMs) with multiple match states that describe local structural information without exploiting explicit structure predictions. Parameters for such models have been estimated from a large library of structure-based alignments. We show that (i) on remote homologs, MUMMALS achieves statistically best accuracy among several leading aligners, such as ProbCons, MAFFT and MUSCLE, albeit the average improvement is small, in the order of several percent; (ii) a large collection (>10 000) of automatically computed pairwise structure alignments of divergent protein domains is superior to smaller but carefully curated datasets for estimation of alignment parameters and performance tests; (iii) reference-independent evaluation of alignment quality using sequence alignment-dependent structure superpositions correlates well with reference-dependent evaluation that compares sequence-based alignments to structure-based reference alignments. The MUMMALS web server is available at: http://prodata.swmed.edu/mummals PDF
We developed PROMALS, a multiple alignment method that shows promising results for protein homologs with sequence identity below 10%, aligning close to half of the amino acid residues correctly on average. This is about three times more accurate than traditional pairwise sequence alignment methods. PROMALS algorithm derives its strength from several sources: (i) sequence database searches to retrieve additional homologs; (ii) accurate secondary structure prediction; (iii) a hidden Markov model that uses a novel combined scoring of amino acids and secondary structures; (iv) probabilistic consistency-based scoring applied to progressive alignment of profiles. Compared to the best alignment methods that do not use secondary structure prediction and database searches (e.g. MUMMALS, ProbCons and MAFFT), PROMALS is up to 30% more accurate, with improvement being most prominent for highly divergent homologs. Compared to SPEM and HHalign, which also employ database searches and secondary structure prediction, PROMALS shows an accuracy improvement of several percent. The PROMALS web server is available at: http://prodata.swmed.edu/promals PDF1 PDF2
The rapidly increasing database of spatial structures is a valuable source to improve alignment quality. We explore the use of 3D structural information to guide sequence alignments constructed by our MSA program PROMALS. The resulting tool, PROMALS3D, automatically identifies homologs with known 3D structures for the input sequences, derives structural constraints through structure-based alignments and combines them with sequence constraints to construct consistency-based multiple sequence alignments. The output is a consensus alignment that brings together sequence and structural information about input proteins and their homologs. PROMALS3D can also align sequences of multiple input structures, with the output representing a multiple structure-based alignment refined in combination with sequence constraints. The advantage of PROMALS3D is that it gives researchers an easy way to produce high-quality alignments consistent with both sequences and structures of proteins. PROMALS3D outperforms a number of existing methods for constructing multiple sequence or structural alignments using both reference-dependent and reference-independent evaluation methods. The PROMALS3D web server is available at: http://prodata.swmed.edu/promals3d PDF1 PDF2
Although pairwise alignment construction has been extensively researched, alignments are still not sufficiently accurate for sequences with low similarity. Automatic aligners such as PROMALS frequently misalign alignment blocks by a few residues. Better alignment solutions can be found among a limited set of local shifts of alignment blocks (moving residues in the query relative to the template). This observation motivated us to develop a pairwise alignment refinement method, SFESA, which generates candidate alignment variants for each alignment block by shifting the query region. We developed a scoring function to judge whether an alignment variant is likely to be more accurate than the original alignment. Our scoring function combines a profile-based sequence score and a novel structural contact-based score derived from residue contacts in template. This combined score was often able to select the best alignment solution among a set of candidates and lead to overall increase in alignment accuracy. Our approach improves alignments generated by a number of methods such as PROMALS, HHpred and CNFpred on several benchmarks that include both reference-dependent and reference-independent assessment. The pairwise alignment refinement tool SFESA is available as an online server at: http://prodata.swmed.edu/sfesa/ PDF
3D structure alignment up
Although plethora of methods for structure alignments exist, the problem of finding equivalent residues in weakly similar structures is not solved. Many factors are considered when experts make manual 3D superpositions and alignments derived from them. Spatial proximity is not enough to produce biologically meaningful alignments. In our algorithm, we are trying to emulate an expert, and to combine superposition strategies with intramolecular contact-based approaches. We try to maximize the number of superimposed residues under the constraints of matching H-bond patterns and side-chain orientations in β-sheets, plus a few key contacts between β-strands and α-helices.
Statistics of protein comparison up
Quantification of statistical significance is essential for the interpretation of protein similarity. To address this, we work on statistical model for sequence and structure comparison.
Comparison of multiple protein sequence alignments (MSA) reveals unexpected evolutionary relations between protein families and leads to exciting predictions of spatial structure and function. The power of MSA comparison critically depends on the quality of statistical model used to rank the similarities found in a database search, so that biologically relevant relationships are discriminated from spurious connections. We developed an accurate statistical description of MSA comparison that does not originate from conventional models of single sequence comparison and captures essential features of protein families. As a final result, we compute E-values for the similarity between any two MSA using a mathematical function that depends on MSA lengths and sequence diversity. To develop these estimates of statistical significance, we first establish a procedure for generating realistic alignment decoys that reproduce natural patterns of sequence conservation dictated by protein secondary structure. Second, since similarity scores between these alignments do not follow the classic Gumbel extreme value distribution, we propose a novel distribution, which we call power-EVD that yields statistically perfect agreement with the data. The probability density function of pEVD is:
where x is the score (random variable), m and s are location and scale parameters, α, β are shape parameters and C is a normalization constant. The four parameters of this distribution depend on sequence length and number of sequences in a profile. Third, we apply this random model to database searches and show that it surpasses conventional models in the accuracy of detecting remote protein similarities. PDF
Profile-based analysis of multiple sequence alignments (MSA) allows for accurate comparison of protein families. We address the problems of detecting statistically confident dissimilarities between (1) MSA position and a set of predicted residue frequencies, and (2) between two MSA positions. These problems are important for (i) evaluation and optimization of methods predicting residue occurrence at protein positions; (ii) detection of potentially misaligned regions in automatically produced alignments and their further refinement; and (iii) detection of sites that determine functional or structural specificity in two related families. For problems (1) and (2), we propose analytical estimates of P-value and apply them to the detection of significant positional dissimilarities in various experimental situations. (a) We compare structure-based predictions of residue propensities at a protein position to the actual residue frequencies in the MSA of homologs. (b) We evaluate our method by the ability to detect erroneous position matches produced by an automatic sequence aligner. (c) We compare MSA positions that correspond to residues aligned by automatic structure aligners. (d) We compare MSA positions that are aligned by high-quality manual superposition of structures. Detected dissimilarities reveal shortcomings of the automatic methods for residue frequency prediction and alignment construction. For the high-quality structural alignments, the dissimilarities suggest sites of potential functional or structural importance. The proposed computational method is of significant potential value for the analysis of protein families. PDF
A random model for protein structure comparison was developed. Novelty of the model is threefold. First, a sample of random structure comparisons is restricted to molecules of the same size and shape as the superposition of interest. Second, careful selection of the sample and accurate modeling of shape allows approximation of the root mean square deviation (RMSD) distribution of random comparisons with a Nakagami probability density function:
where x is RMS deviation of coordinates in a superposition of two structures (random variable), k and s are parameters of the distribution and Γ is Euler Gamma function.
Third, through convolution, a second probability density function is obtained that describes the coordinate difference vector projections underlying the random distribution of RMSD. This last feature allows sampling random distributions of not only RMSD, but also any similarity score that depends on difference vector projections, such as GDT–TS score, TM score, and LiveBench 3D score. Probabilities estimated from the method correlate well with common measures of structural similarity, such as the Dali Z-score and the GDT–TS score. As a result, the p-value for a given superposition can be calculated using simple formulae depending on RMSD, radius of gyration, and thinnest molecular dimension. In addition to scoring structural similarity, p-values computed by this method can be applied to evaluation of homology modeling techniques, providing a statistically sound alternative to scores used in reference-independent evaluation of alignment quality. PDF
Ancestral sequence reconstruction up
Modern-day proteins were selected during long evolutionary history as descendants of ancient life forms. In silico reconstruction of such ancestral protein sequences facilitates our understanding of evolutionary processes, protein classification and biological function. Additionally, reconstructed ancestral protein sequences could serve to fill in sequence space thus aiding remote homology inference. We developed ANCESCON, a package for distance-based phylogenetic inference and reconstruction of ancestral protein sequences that takes into account the observed variation of evolutionary rates between positions that more precisely describes the evolution of protein families. To improve the accuracy of evolutionary distance estimation and ancestral sequence reconstruction, two approaches are proposed to estimate position-specific evolutionary rates. Comparisons show that at large evolutionary distances our method gives more accurate ancestral sequence reconstruction than PAML, PHYLIP and PAUP*. We apply the reconstructed ancestral sequences to homology inference and functional site prediction. We show that the usage of hypothetical ancestors together with the present day sequences improves profile-based sequence similarity searches; and that ancestral sequence reconstruction methods can be used to predict positions with functional specificity. As a computational tool to reconstruct ancestral protein sequences from a given multiple sequence alignment, ANCESCON shows high accuracy in tests and helps detection of remote homologs and prediction of functional sites. ANCESCON is freely available for non-commercial use. Pre-compiled versions for several platforms can be downloaded from http://prodata.swmed.edu/download/pub/ANCESCON and the web server is set up here. PDF
Phylogenetic trees up
The reliable reconstruction of tree topology from a set of homologous sequences is one of the main goals in the study of molecular evolution. If consistent estimators of distances from a multiple sequence alignment are known, the distance method is attractive because the tree reconstruction is consistent. To obtain a distance estimate d, the observed proportion of differences p (p-distance) is usually "corrected" for multiple and back substitutions by means of a functional relationship d = f(p). We derived conditions under which this correction of p-distances will not alter the selection of the tree topology are specified. When these conditions are not fulfilled the selection of the tree topology may depend on the correction function applied. A novel method which includes estimates of distances not only between sequence pairs, but between triplets, quadruplets, etc., was proposed to strengthen the proper selection of correction function and tree topology. PDF
Prediction of structural cores in proteins up
The structures of homologous proteins are generally better conserved than their sequences. This phenomenon is demonstrated by the prevalence of structurally conserved regions (SCRs) even in highly divergent protein families. Defining SCRs requires the comparison of two or more homologous structures and is affected by their availability and divergence, and our ability to deduce structurally equivalent positions among them. In the absence of multiple homologous structures, it is necessary to predict SCRs of a protein using information from only a set of homologous sequences and (if available) a single structure. Accurate SCR predictions can benefit homology modelling and sequence alignment. Using pairwise DaliLite alignments among a set of homologous structures, we devised a simple measure of structural conservation, termed structural conservation index (SCI). SCI was used to distinguish SCRs from non-SCRs. A database of SCRs was compiled from 386 SCOP superfamilies containing 6489 protein domains. Artificial neural networks were then trained to predict SCRs with various features deduced from a single structure and homologous sequences. Assessment of the predictions via a 5-fold cross-validation method revealed that predictions based on features derived from a single structure perform similarly to ones based on homologous sequences, while combining sequence and structural features was optimal in terms of accuracy (0.755) and Matthews correlation coefficient (0.476). These results suggest that even without information from multiple structures, it is still possible to effectively predict SCRs for a protein. Finally, inspection of the structures with the worst predictions pinpoints difficulties in SCR definitions. The SCR database and the prediction server can be found here: http://prodata.swmed.edu/SCR/ PDF
Prediction of nuclear export signals up
Classical nuclear export signals (NESs) are short cognate peptides that direct proteins out of the nucleus via the CRM1-mediated export pathway. CRM1 regulates the localization of hundreds of macromolecules involved in various cellular functions and diseases. Due to the diverse and complex nature of NESs, reliable prediction of the signal remains a challenge despite several attempts made in the last decade. In a collaboration with the Chook Lab, we developed a new NES predictor, LocNES. LocNES scans query proteins for NES consensus-fitting peptides and assigns these peptides probability scores using Support Vector Machine model, whose feature set includes amino acid sequence, disorder propensity, and the rank of position-specific scoring matrix score. LocNES demonstrates both higher sensitivity and precision over existing NES prediction tools upon comparative analysis using experimentally identified NESs. LocNES is freely available at http://prodata.swmed.edu/LocNES/ PDF
In light of the currently resolved crystal structures of CRM1-NES peptides with diverse classes, we have also been working on developing a structure-based predictor to find a breakthrough for distinguishing r eal NESs and false positives. By combining the sequence-based and structure-based approaches, we analyzed the structural prerequisites for CRM1-dependent NESs, i.e., accessibility (by locating disordered/orde red regions), adapting conformation (by predicting secondary structures), and the stability at the binding site (by applying structure-based modeling to calculate binding energies). For a subset of validated NES peptides, the predicted binding energies correlate well to the experimental binding affinities, and we can distinguish the real NES motifs and false positives which both match NES consensus patterns. We a re continuously optimizing our program by the iterative process of energy prediction and experimental validation. PDF
Better defined NES should also allow meaningful mapping of cancer-related mutation positions, leading to plausible explanations for the relationship between nuclear export and disease. We extracted possible N ES candidate regions from the cancer-related human reference proteome and scored the sequence segments for reliability as NES. The confidently identified NES candidate motifs were checked for overlap with can cer-related mutation positions annotated in the COSMIC database. Among the ∼700 cancer-related sequences in the COSMIC Cancer Gene Census, 178 sequences are predicted to have possible NES motifs containing c ancer-related mutations at their key positions. These lists are organized into our database (pCRM1exportome), which is freely available at http://prodata.swmed. edu/pCRM1exportome/. PDF
Prediction of functional sites up
A number of methods have been developed to predict functional specificity determinants in protein families based on sequence information. Most of these methods rely on pre-defined functional subgroups. Manual subgroup definition is difficult because of the limited number of experimentally characterized subfamilies with differing specificity, while automatic subgroup partitioning using computational tools is a non-trivial task and does not always yield ideal results. We propose a new approach SPEL (specificity positions by evolutionary likelihood) to detect positions that are likely to be functional specificity determinants. SPEL, which does not require subgroup definition, takes a multiple sequence alignment of a protein family as the only input, and assigns a P-value to every position in the alignment. Positions with low P-values are likely to be important for functional specificity. An evolutionary tree is reconstructed during the calculation, and P-value estimation is based on a random model that involves evolutionary simulations. Evolutionary log-likelihood is chosen as a measure of amino acid distribution at a position. To illustrate the performance of the method, we carried out a detailed analysis of two protein families (LacI/PurR and G protein alpha subunit), and compared our method with two existing methods (evolutionary trace and mutual information based). All three methods were also compared on a set of protein families with known ligand-bound structures. SPEL is freely available for non-commercial use. Its pre-compiled versions for several platforms and alignments used in this work are available at http://prodata.swmed.edu/download/pub/SPEL PDF
Protein fold and structure prediction up
We worked on development of approaches to side-chain modeling, sequence design and local structure prediction.
Modeling side-chain conformations on a fixed protein backbone has a wide application in structure prediction and molecular design. Each effort in this field requires decisions about a rotamer set, scoring function, and search strategy. We have developed a new and simple scoring function, which operates on side-chain rotamers and consists of the following energy terms: contact surface, volume overlap, backbone dependency, electrostatic interactions, and desolvation energy. The weights of these energy terms were optimized to achieve the minimal average root mean square (rms) deviation between the lowest energy rotamer and real side-chain conformation on a training set of high-resolution protein structures. In the course of optimization, for every residue, its side chain was replaced by varying rotamers, whereas conformations for all other residues were kept as they appeared in the crystal structure. We obtained prediction accuracy of 90.4% for χ1, 78.3% for χ1+2, and 1.18 A overall rms deviation. Furthermore, the derived scoring function combined with a Monte Carlo search algorithm was used to place all side chains onto a protein backbone simultaneously. The average prediction accuracy was 87.9% for χ1, 73.2% for χ1+2, and 1.34 A rms deviation for 30 protein structures. Our approach was compared with available side-chain construction methods and showed improvement over the best among them: 4.4% for χ1, 4.7% for χ1+2, and 0.21 A for rms deviation. We hypothesize that the scoring function instead of the search strategy is the main obstacle in side-chain modeling. Additionally, we show that a more detailed rotamer library is expected to increase χ1+2 prediction accuracy but may have little effect on χ1 prediction accuracy. PDF
We have developed an effective scoring function for protein design. The atomic solvation parameters, together with the weights of energy terms, were optimized so that residues corresponding to the native sequence were predicted with low energy in the training set of 28 protein structures. The solvation energy of non-hydrogen-bonded hydrophilic atoms was considered separately and expressed in a nonlinear way. As a result, our scoring function predicted native residues as the most favorable in 59% of the total positions in 28 proteins. We then tested the scoring function by comparing the predicted stability changes for 103 T4 lysozyme mutants with the experimental values. The correlation coefficients were 0.77 for surface mutations and 0.71 for all mutations. Finally, the scoring function combined with Monte Carlo simulation was used to predict favorable sequences on a fixed backbone. The designed sequences were similar to the natural sequences of the family to which the template structure belonged. The profile of the designed sequences was helpful for identification of remote homologues of the native sequence. PDF
We studied the effects of various factors in representing and combining evolutionary and structural information for local protein structural prediction based on fragment selection. We prepare databases of fragments from a set of non-redundant protein domains. For each fragment, evolutionary information is derived from homologous sequences and represented as estimated effective counts and frequencies of amino acids (evolutionary frequencies) at each position. Position-specific amino acid preferences called structural frequencies are derived from statistical analysis of discrete local structural environments in database structures. Our method for local structure prediction is based on ranking and selecting database fragments that are most similar to a target fragment. Using secondary structure type as a local structural property, we test our method in a number of settings. The major findings are: (1) the COMPASS-type scoring function for fragment similarity comparison gives better prediction accuracy than three other tested scoring functions for profile-profile comparison. We show that the COMPASS-type scoring function can be derived both in the probabilistic framework and in the framework of statistical potentials. (2) Using the evolutionary frequencies of database fragments gives better prediction accuracy than using structural frequencies. (3) Finer definition of local environments, such as including more side-chain solvent accessibility classes and considering the backbone conformations of neighboring residues, gives increasingly better prediction accuracy using structural frequencies. (4) Combining evolutionary and structural frequencies of database fragments, either in a linear fashion or using a pseudocount mixture formula, results in improvement of prediction accuracy. Combination at the log-odds score level is not as effective as combination at the frequency level. This suggests that there might be better ways of combining sequence and structural information than the commonly used linear combination of log-odds scores. Our method of fragment selection and frequency combination gives reasonable results of secondary structure prediction tested on 56 CASP5 targets (average SOV score 0.77), suggesting that it is a valid method for local protein structure prediction. Mixture of predicted structural frequencies and evolutionary frequencies improve the quality of local profile-to-profile alignment by COMPASS. PDF
Other work in this area was aimed at predicting structures from various protein families.
The complete sequence of the male-specific region of the human Y chromosome (MSY) has been determined recently; however, detailed characterization for many of its encoded proteins still remains to be done. We applied state-of-the-art protein structure prediction methods to all 27 distinct MSY-encoded proteins to provide better understanding of their biological functions and their mechanisms of action at the molecular level. The results of such large-scale structure-functional annotation provide a comprehensive view of the MSY proteome, shedding light on MSY-related processes. We found that, in total, at least 60 domains are encoded by 27 distinct MSY genes, of which 42 (70%) were reliably mapped to currently known structures. The most challenging predictions include the unexpected but confident 3D structure assignments for three domains identified here encoded by the USP9Y, UTY, and BPY2 genes. The domains with unknown 3D structures that are not predictable with currently available theoretical methods are established as primary targets for crystallographic or NMR studies. The data presented here set up the basis for additional scientific discoveries in human biology of the Y chromosome, which plays a fundamental role in sex determination. PDF
The GGDEF domain is detected in many prokaryotic proteins, most of which are of unknown function. Several bacteria carry 12-22 different GGDEF homologues in their genomes. Conducting extensive profile-based searches, we detect statistically supported sequence similarity between GGDEF domain and adenylyl cyclase catalytic domain. From this homology, we deduce that the prokaryotic GGDEF domain is a regulatory enzyme involved in nucleotide cyclization, with the fold similar to that of the eukaryotic cyclase catalytic domain. This prediction correlates with the functional information available on two GGDEF-containing proteins, namely diguanylate cyclase and phosphodiesterase A of Acetobacter xylinum, both of which regulate the turnover of cyclic diguanosine monophosphate. Domain architecture analysis shows that GGDEF is typically present in multidomain proteins containing regulatory domains of signaling pathways or protein-protein interaction modules. Evolutionary tree analysis indicates that GGDEF/cyclase superfamily forms a large diversified cluster of orthologous proteins present in bacteria, archaea, and eukaryotes. Structure of GGDEF domain has been since solved and confirmed our prediction. PDF
γ-Glultamylcysteine synthetase (γ-GCS) catalyzes the first step in the de novo biosynthesis of glutathione. In trypanosomes, glutathione is conjugated to spermidine to form a unique cofactor termed trypanothione, an essential cofactor for the maintenance of redox balance in the cell. Using extensive similarity searches and sequence motif analysis we detected homology between γ-GCS and glutamine synthetase (GS), allowing these proteins to be unified into a superfamily of carboxylate-amine/ammonia ligases. The structure of γ-GCS, which was previously poorly understood, was modeled using the known structure of GS. Two metal-binding sites, each ligated by three conserved active site residues (n1: Glu-55, Glu-93, Glu-100; and n2: Glu-53, Gln-321, and Glu-489), are predicted to form the catalytic center of the active site, where the n1 site is expected to bind free metal and the n2 site to interact with MgATP. To elucidate the roles of the metals and their ligands in catalysis, these six residues were mutated to alanine in the Trypanosoma brucei enzyme. All mutations caused a substantial loss of activity. Most notably, E93A was able to catalyze the l-Glu-dependent ATP hydrolysis but not the peptide bond ligation, suggesting that the n1 metal plays an important role in positioning l-Glu for the reaction chemistry. The apparent Km values for ATP were increased for both the E489A and Q321A mutant enzymes, consistent with a role for the n2 metal in ATP binding and phosphoryl transfer. Furthermore, the apparent K(d) values for activation of E489A and Q321A by free Mg2+ increased. Finally, substitution of Mn2+ for Mg2+ in the reaction rescued the catalytic deficits caused by both mutations, demonstrating that the nature of the metal ligands plays an important role in metal specificity. PDF
Two different type II topoisomerases are known in bacteria. DNA gyrase (Gyr) introduces negative supercoils into DNA. Topoisomerase IV (Par) relaxes DNA supercoils. GyrA and ParC subunits of bacterial type II topoisomerases are involved in breakage and reunion of DNA. The spatial structure of the C-terminal fragment in GyrA/ParC is not available. We infer homology between the C-terminal domain of GyrA/ParC and a regulator of chromosome condensation (RCC1), a eukaryotic protein that functions as a guanine-nucleotide-exchange factor for the nuclear G protein Ran. This homology, complemented by detection of 6 sequence repeats with 4 predicted beta-strands each in GyrA/ParC sequences, allows us to predict that the GyrA/ParC C-terminal domain folds into a 6-bladed beta-propeller. The prediction rationalizes available experimental data and sheds light on the spatial properties of the largest topoisomerase domain that lacks structural information. PDF
Our group teamed up with the Baker group to predict structures in CASP8. The Baker group worked on model refinement and de novo predictions, and we contributed template identification, alignments and manual curation of models. Aggressive sampling and all-atom refinement were carried out for nearly all targets. A combination of alignment methodologies was used to generate starting models from a range of templates, and the models were then subjected to Rosetta all atom refinement. For the 64 domains with readily identified templates, the best submitted model was better than the best alignment to the best template in the Protein Data Bank for 24 cases, and improved over the best starting model for 43 cases. For 13 targets where only very distant sequence relationships to proteins of known structure were detected, models were generated using the Rosetta de novo structure prediction methodology followed by all-atom refinement; in several cases the submitted models were better than those based on the available templates. Of the 12 refinement challenges, the best submitted model improved on the starting model in seven cases. These improvements over the starting template-based models and refinement tests demonstrate the power of Rosetta structure refinement in improving model accuracy. As an interesting example, T0487 was the largest structured protein ever evaluated in CASP (685 residues). The overall strategy for predicting the structure of this protein involved refining each domain separately to test different alignment variants, assembling the best individually refined domains onto the full-length 2f8s template, and refining the complete model again. Out of the five domains comprising this target, we did very well on T0487 domain 4 (model01 GDT-TS is 79.2 and GDT-TS Z-score is 4.50). Two template sequences (1yvu/2f8s and 1u04) that correspond to the full-length target were identified with BLAST, while additional template sequences related to individual domains were identified with PSI-BLAST (1w9h includes domain 1 and 5, and 1r4k, 1si2, and 1r6z include domain 3). A structure based alignment of identified templates displayed sequence regions with an inconsistent hydrophobicity profile. Because the hydrophobicity patterns of a template with lower resolution (2f8s) agreed better with the PROMALS target family alignment than the template with the best resolution (1u04), the lower resolution template (2f8s) was chosen. The fourth domain of T0487 (res. 177–265) forms an SH3-like barrel known as a PAZ domain. Of the identified templates, the individual PAZ domain of human eIF2c1 bound to a 3' siRNA-like deoxynucleotide overhang (1si2) represented the closest non-NMR template sequence. A PROMALS3D multiple alignment of this domain to all templates was adjusted manually to preserve hydrophobicity patterns and conserved residues in the 3' overhang-binding site. Two templates were chosen for initial refinement: (1) the individual PAZ domain template 1si2 and (2) a hybrid of this template substituting the first 19 residues from the full-length template 2f8s. Various alignment variants were tested by model refinement, with the lowest energy model corresponding to one alignment variant with the hybrid template. After assembly of this alignment variant into the full-length template 2f8s, subsequent refinement produced the excellent model shown in the right panel of the Figure above. PDF
Assessment of structure prediction up
Our group classified targets and assessed fold recognition category in CASP5, and we introduced a few new twists into model evaluations. Prediction models were evaluated by using six different structural measures and four different alignment measures, and these scores were compared to those assigned manually over a diverse subset of target domains. Scores were combined to compare overall performance of participating groups and to estimate rank significance. The best assessment methodology was the one that combined all the measures, as it correlated best with manual analysis of predictions. The methods used by a few groups outperformed all other methods in terms of the evaluated criteria and could be considered state-of-the-art in structure prediction. We also compared the results of manual groups to those of automatic servers evaluated in parallel by CAFASP, showing that the top performing automated server structure predictions approached those of the best manual predictors. Recently, we worked out a web interface to display model evaluation results and to show target analysis using CASP8 targets and server predictions. PDF
Our groups was asked to assess Free Modeling Category predictions in CASP9. We applied a similar comprehensive score that combines ten different structure and sequence measurements and compared it with manual assessment on a diverse subset. A new QCS score was also developed to mimic manual decisions by capturing both global and local structural features. To rank the overall performance of participating groups, prediction scores were rescaled to Z-scores and summed up to produce a single value. We observed a slight overall increase in performance compared to previous years, despite the increased difficulty of targets. Some models were better than closest structure templates. Notably, a server prediction model for a single target (T0581) improved significantly over the closest structure template (44% GDT increase). This accomplishment represents the "winner" of the CASP9 FM category. A number of human expert groups submitted slight variations of this model, highlighting a trend for human experts to act as "meta predictors" by correctly selecting among models produced by the top-performing automated servers. The detailed assessment results can be viewed at our website http://prodata.swmed.edu/CASP9/ PDF
Manual inspection has been applied to and is well accepted for assessing critical assessment of protein structure prediction (CASP) free modeling (FM) category predictions over the years. Such manual assessment requires expertise and significant time investment, yet has the problems of being subjective and unable to differentiate models of similar quality. It is beneficial to incorporate the ideas behind manual inspection to an automatic score system, which could provide objective and reproducible assessment of structure models. Inspired by our experience in CASP9 FM category assessment, we developed an automatic superimposition independent method named Quality Control Score (QCS) for structure prediction assessment. QCS captures both global and local structural features, with emphasis on global topology. We applied this method to all FM targets from CASP9, and overall the results showed the best agreement with Manual Inspection Scores among automatic prediction assessment methods previously applied in CASPs, such as Global Distance Test Total Score (GDT_TS) and Contact Score (CS). As one of the important components to guide our assessment of CASP9 FM category predictions, this method correlates well with other scoring methods and yet is able to reveal good-quality models that are missed by GDT_TS. The scripts for QCS calculation is available at http://prodata.swmed.edu/QCS/ PDF
Accurate and sensitive performance evaluation is crucial for both effective development of better structure prediction methods based on sequence similarity, and for the comparative analysis of existing methods. Up to date, there has been no satisfactory comprehensive evaluation method that (i) is based on a large and statistically unbiased set of proteins with clearly defined relationships; and (ii) covers all performance aspects of sequence-based structure predictors, such as sensitivity and specificity, alignment accuracy and coverage, and structure template quality. With the aim of designing such a method, we (i) select a statistically balanced set of divergent protein domains from SCOP, and define similarity relationships for the majority of these domains by complementing the best of information available in SCOP with a rigorous SVM-based algorithm; and (ii) develop protocols for the assessment of similarity detection and alignment quality from several complementary perspectives. The evaluation of similarity detection is based on ROC-like curves and includes several complementary approaches to the definition of true/false positives. Reference-dependent approaches use the 'gold standard' of pre-defined domain relationships and structure-based alignments. Reference-independent approaches assess the quality of structural match predicted by the sequence alignment, with respect to the whole domain length (global mode) or to the aligned region only (local mode). Similarly, the evaluation of alignment quality includes several reference-dependent and -independent measures, in global and local modes. As an illustration, we use our benchmark to compare the performance of several methods for the detection of remote sequence similarities, and show that different aspects of evaluation reveal different properties of the evaluated methods, highlighting their advantages, weaknesses, and potential for further development. The presented benchmark provides a new tool for a statistically unbiased assessment of methods for remote sequence similarity detection, from various complementary perspectives. This tool should be useful both for users choosing the best method for a given purpose, and for developers designing new, more powerful methods. The benchmark set, reference alignments, and evaluation codes can be downloaded from http://prodata.swmed.edu/download/pub/evaluation/ PDF
Recent improvement in homology-based structure modeling emphasizes the importance of sensitive evaluation measures that help identify and correct modest distortions in models compared to the target structures. GDT_TS, otherwise a very powerful and effective measure for model evaluation, is still insensitive to and can even reward such distortions, as observed for remote homology modeling in the latest Comparative Assessment of Structure Prediction, CASP8. We develop a new measure that balances GDT_TS reward for the closeness of equivalent model and target residues ("attraction" term) with the penalty for the closeness of non-equivalent residues ("repulsion" term). Compared to GDT_TS, the resulting score, TR, is much more sensitive to structure compression both in real remote homologs and in CASP models. TR is correlated yet different from other measures of structure similarity. The largest difference from GDT_TS is observed in models of mid-range quality based on remote homology modeling.PDF
Web-servers for protein analysis up
Computational sequence analysis, that is, prediction of local sequence properties, homologs, spatial structure and function from the sequence of a protein, offers an efficient way to obtain needed information about proteins under study. Since reliable prediction is usually based on the consensus of many computer programs, meta-severs have been developed to fit such needs. Most meta-servers focus on one aspect of sequence analysis, while others incorporate more information. However, as predictions of local sequence properties, three-dimensional structure and function are usually intertwined, it is beneficial to address them together. We developed a MEta-Server for protein Sequence Analysis (MESSA) to facilitate comprehensive protein sequence analysis and gather structural and functional predictions for a protein of interest. For an input sequence, the server exploits a number of select tools to predict local sequence properties, such as secondary structure, structurally disordered regions, coiled coils, signal peptides and transmembrane helices; detect homologous proteins and assign the query to a protein family; identify three-dimensional structure templates and generate structure models; and provide predictive statements about the protein's function, including functional annotations, Gene Ontology terms, enzyme classification and possible functionally associated proteins. MESSA is free for non-commercial use at http://prodata.swmed.edu/MESSA/. PDF
The size of the protein sequence database has been exponentially increasing due to advances in genome sequencing. However, experimentally characterized proteins only constitute a small portion of the database, such that the majority of sequences have been annotated by computational approaches. Current automatic annotation pipelines inevitably introduce errors, making the annotations unreliable. Instead of such error-prone automatic annotations, functional interpretation should rely on annotations of 'reference proteins' that have been experimentally characterized or manually curated. The Seq2Ref server uses BLAST to detect proteins homologous to a query sequence and identifies the reference proteins among them. Seq2Ref then reports publications with experimental characterizations of the identified reference proteins that might be relevant to the query. Furthermore, a plurality-based rating system is developed to evaluate the homologous relationships and rank the reference proteins by their relevance to the query. The reference proteins detected by our server will lend insight into proteins of unknown function and provide extensive information to develop in-depth understanding of uncharacterized proteins. Seq2Ref is free for non-commercial use at http://prodata.swmed.edu/Seq2Ref/. PDF
One approach to infer functions of new proteins from their homologs utilizes visualization of an all-against-all pairwise similarity network (A2ApsN) that exploits the speed of BLAST and avoids the complexity of multiple sequence alignment. However, identifying functions of the protein clusters in A2ApsN is never trivial, due to a lack of linking characterized proteins to their relevant information in current software packages. Given the database errors introduced by automatic annotation transfer, functional deduction should be made from proteins with experimental studies, i.e., 'reference proteins'. Here, we present a web server, termed Pclust, which provides a user-friendly interface to visualize protein similarity network, placing emphasis on such 'reference proteins' and providing access to their full information in source databases, e.g., articles in PubMed. The identification of 'reference proteins' and the ease of cross-database linkage will facilitate understanding the functions of protein clusters in the network, thus promoting interpretation of proteins of interest. Pclust is free for non-commercial use at http://prodata.swmed.edu/Pclust/. PDF
Online Mendelian Inheritance in Man (OMIM) is a manually curated compendium of human genetic variants and the corresponding phenotypes, mostly human diseases. Instead of directly documenting the native sequences for gene entries, OMIM links its entries to protein and DNA sequences in other databases. However, because of the existence of gene isoforms and errors in OMIM records, mapping a specific OMIM mutation to its corresponding protein sequence is not trivial. Combining computer programs and extensive manual curation of OMIM full-text descriptions and original literature, we mapped 98% of OMIM amino acid substitutions (AASs) and all SwissProt Variant (SwissVar) disease-related AASs to reference sequences and confidently mapped 99.96% of all AASs to the genomic loci. Based on the results, we developed an online database and interactive web server (M2SG) to (i) retrieve the mapped OMIM and SwissVar variants for a given protein sequence; and (ii) obtain related proteins and mutations for an input disease phenotype. This database will be useful for analyzing sequences, understanding the effect of mutations, identifying important genetic variations and designing experiments on a protein of interest. M2SG is free for non-commercial use at http://prodata.swmed.edu/M2S/. PDF