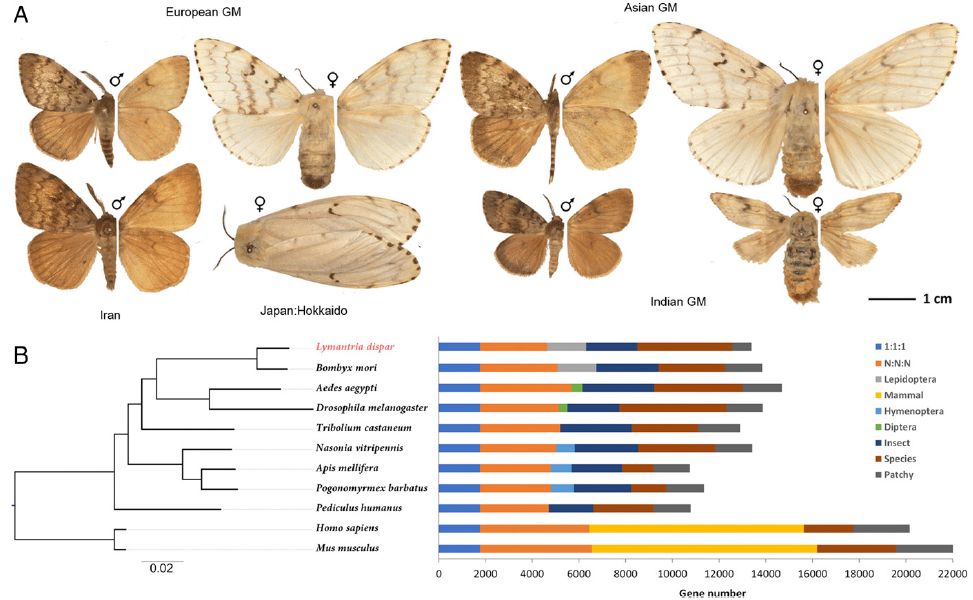
The Gypsy moth as a model organism. (A)
Morphology of Lymantria dispar populations. For each specimen, dorsal (Left) and ventral (Right) sides are shown
and their voucher codes are (left to right, top to bottom): NVG-17104G03, NVG-17104G10, NVG-17104H08, NVG-17105A01, NVG-17104H01, NVG-17105A06, NVG-18028H06, and
NVG-18028H07. See Dataset S1 in the paper for specimen data. (B) Orthology assignment of nine insect and two mammalian genomes. Bars are subdivided to represent
different types of ortholog relationships: "1:1:1" indicates universal single-copy genes present in all species; "Diptera" indicates Dipteran-specific genes,
present in both D. melanogaster and Aedes aegypti; "Hymenoptera" indicates Hymenopteran-specific genes, present in N. vitripennis,
Apis mellifera, and
Pogonomyrmex barbatus genomes; "Insect" indicates all other insect-specific orthologs; "Mammal" indicates mammalian-specific orthologs; "N:N:N" indicates
other universal genes, but absence in a single genome is tolerated; "Patchy" indicates orthologs that are present in at least one insect and one
mammalian genome; "Species" indicates species-specific genes. The phylogeny on the Left is a maximum-likelihood tree of a concatenated alignment of
1,756 single-copy proteins from the 1:1:1 subgroup. The tree was rooted using mammals as the outgroup. CEGMA: these are essential genes and the presence
of them in a genome is used to evaluate the quality of an assembly.