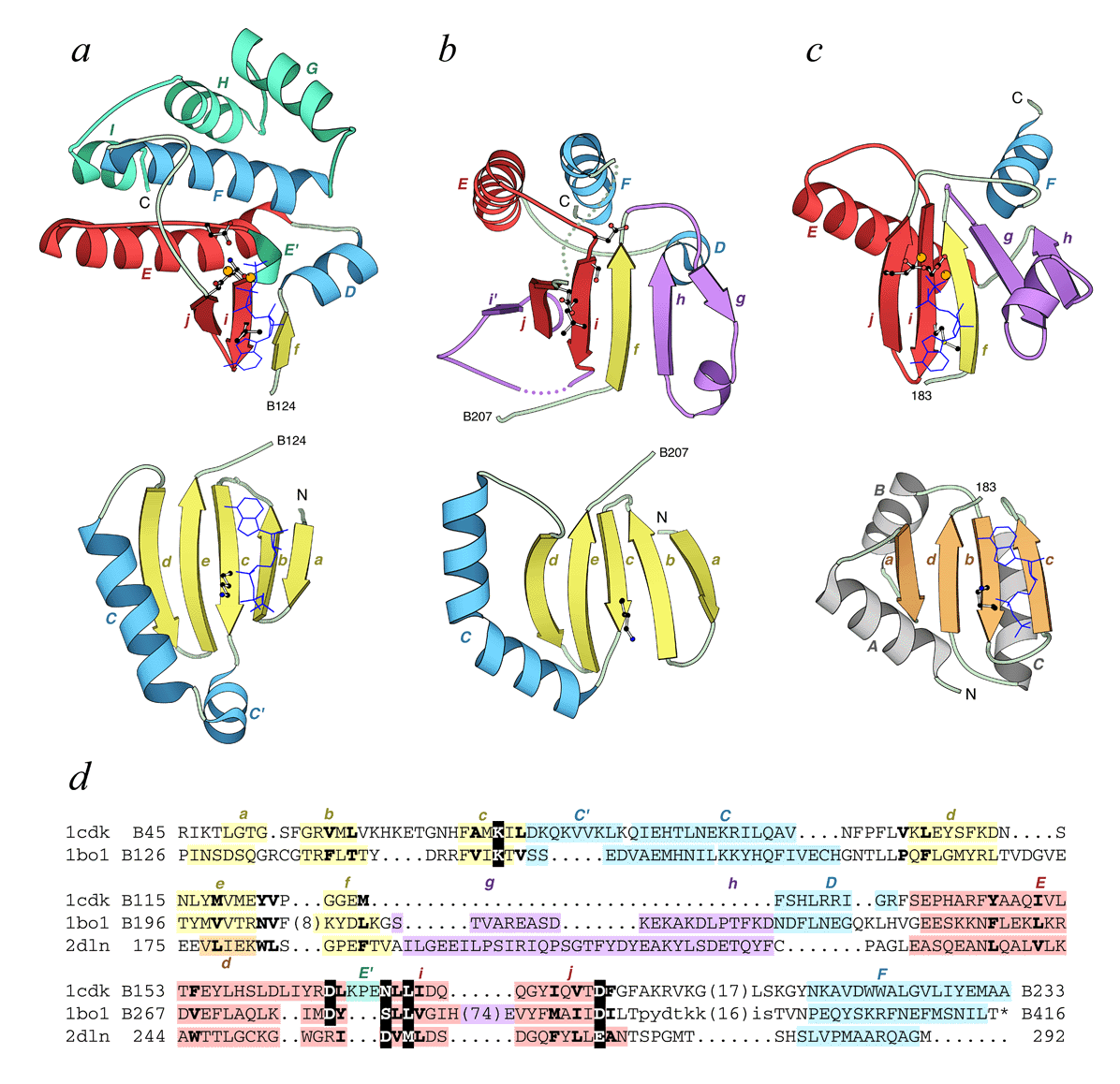
Comparison of kinase and glutathione synthase folds. "Open book" ribbon diagrams of PK, PIPK, SAICAR synthase, and D-ala-D-ala ligase structures are drawn using the program MOLSCRIPT. The N-terminal domain (bottom row) and the C-terminal domain (top row) are separated from each other for the purposes of clarity. The domain interface (ATP-binding cleft) is facing up (open book). The N and C termini of the displayed segment of a protein chain are labeled in roman upper case black letters as well as the numbers of the last and the first residue in the N-terminal and the C-terminal domain, respectively. Secondary structural elements are labeled by color-coded italicized letters: lower case is for β-strands and upper case is for α-helices with letter colors corresponding to the colors of the structural element. The αββ-unit, essential for ATP-binding is shown in red. "Cofactor" is shown in a dark blue wire presentation and is indicated on each domain to mark the binding site. Metal cations are shown as orange balls. Catalytic, ATP and metal-binding residues are shown in a ball-and-stick representation. (a) cAMP-dependent protein kinase in complex with adenylyl imidodiphosphate and Mn2+ (1cdk, residues B46-B298). The additional helices are shown in green. (b) Type IIb phosphatidylinositol phosphate kinase (1bo1, residues B126-B416). (c) SAICAR synthase (1a48, residues 11-272). (d) D-ala-D-ala ligase in complex with ADP and Mg2+ (2dln, residues 95-292). The N-terminal domain of ATP-grasp fold (the "central" domain in the protein) is colored differently to emphasize the fold difference. The additional structural elements in the C-terminal domain (compared to PKs) are shown in purple in (b), (c) and (d). Dots stand for the disordered regions. (e) Structure-based sequence alignment of 1cdk, 1bo1, 1a48, and 2dln. The alignment was constructed using DALI and corrected manually to incorporate additional elements and adjust several regions. Color and letter-coding of structural elements in the sequence is as described for (a), (b), (c) and (d). The chain ID (where available) and the residue number of the first amino acid in each line and the last amino acid in the alignment are indicated. Numbers in parentheses interrupting the sequence indicate the number of residues in insertions not displayed in the alignment. Residues disordered in the structure are shown in lower case letters. Residues that form the conserved hydrophobic core are shown in bold letters. Residues that participate in catalysis, ATP and metal cation binding (displayed in ball-and-stick models in (a), (b), (c) and (d)) and catalysis are shown in white on black.