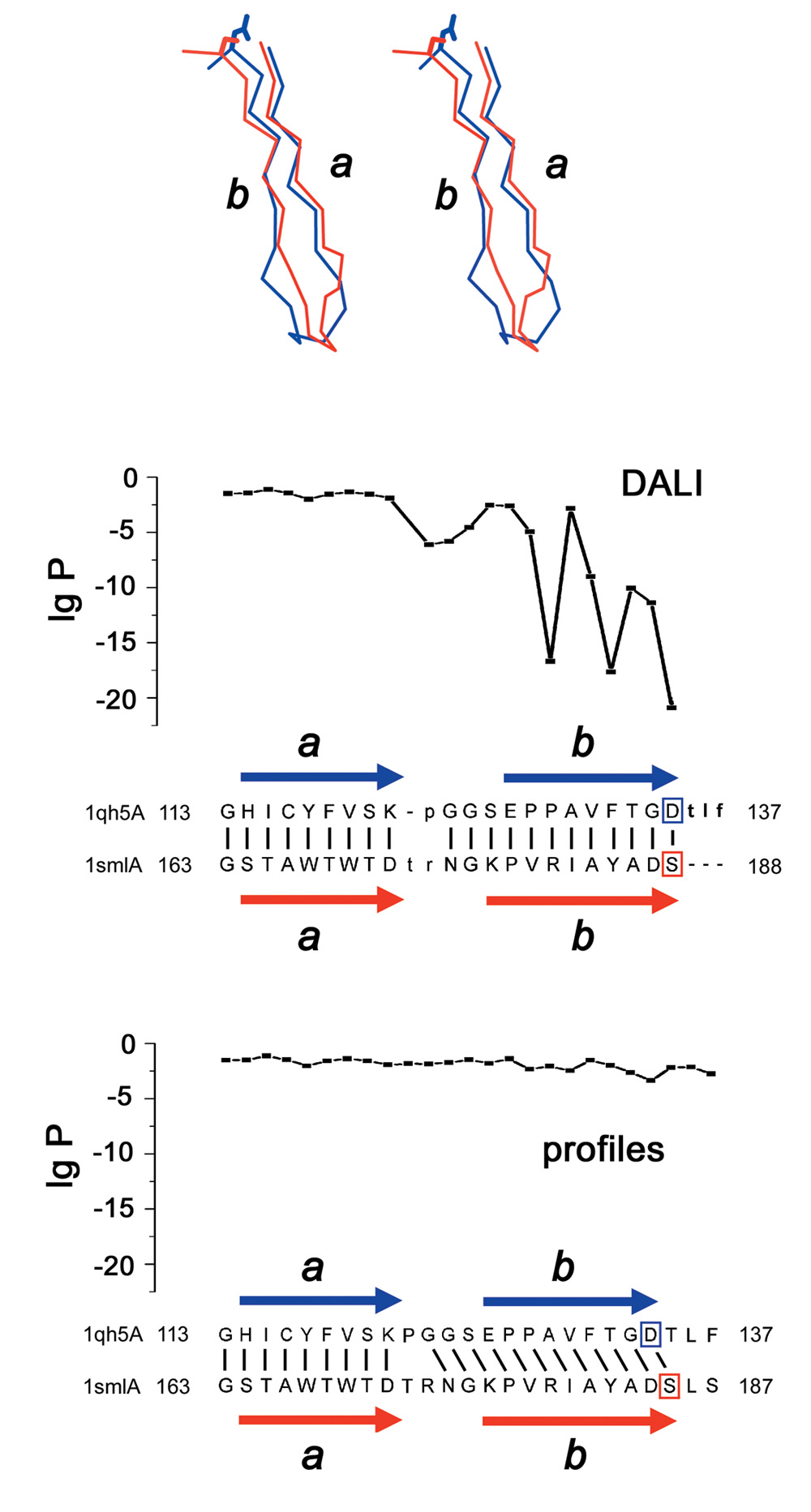
Positional P-values for multiple sequence alignments help revealing structural changes in evolutionary related proteins: Top: Manual alignment of fragments of human glyoxalase II (PDB ID 1qh5A) and bacterial penicillinase (PDB ID 1sml). Strands b are shifted by one residue, so that structurally similar residues correspond to different sequence positions. Bottom: structure alignment by DALI is correct, but low positional P-values reveal contradiction to the sequence similarity, as opposed to higher P-values for the profile-based alignment of the two sequence families. β-strands a and b are shown as arrows, Zn-binding residues are boxed.