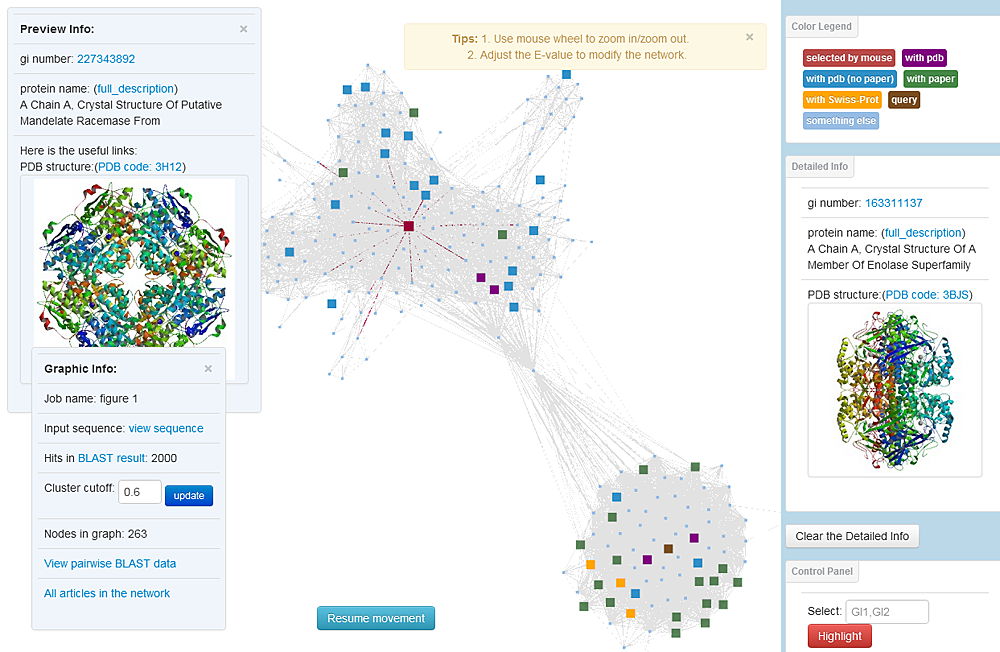
Snapshot of an A2ApsN from Pclust. Protein nodes are colored, hierarchically, brown (input by the user, if applicable), red (selected by mouse), purple (with PDB structures and their articles), green (with PubMed articles), yellow (with Swiss-Prot functional comments), blue (with PDB structures of no article, such as those from structural genomics) and light blue (without any reference, smaller size). A 'preview' panel and a 'detailed Info' panel appear for the protein on which your mouse hovers and your mouse clicks, respectively. Batch selection of proteins is available by inputting the gi numbers (if applicable) separated by commas, and keyword search is available for annotations from the NCBI protein database. By default, Pclust will include the first quarter or 5000 (whichever is smaller) network links (ordered by E-value) and report the corresponding E-value cutoff; a panel to adjust network links by varying the E-value cutoff or the link number is also available. To know more about the control panel, please refer to: http://youtu.be/XLkfEg2jGOc