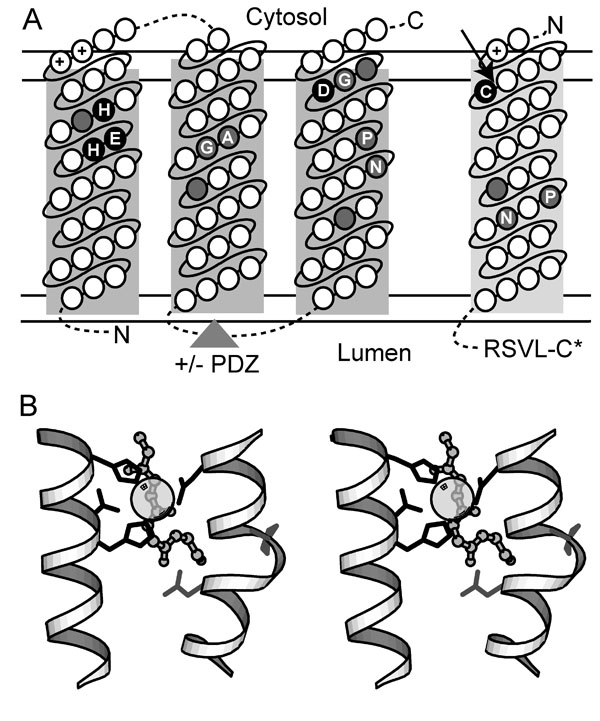
S2P Models. (A) Transmembrane topology of conserved S2P core TMH segments (predicted TMH1-TMH3, boxed in dark gray) and its SREBP substrate (predicted TMH, boxed in light gray) is indicated with the cytosol and lumen labeled above and below the membrane, respectively (bounded by double horizontal lines). Circles located within the boxed segments represent amino acid residues ordered consecutively (connected by solid lines) from the N terminus to the C terminus (labeled accordingly), with various insertions (indicated by dotted lines). The triangle indicates the placement of the PDZ domain present in some family members. Conserved residues required for catalysis are highlighted in black and labeled according to conserved residue type. Positions corresponding to small residue conservations are highlighted in gray and labeled according to conserved residue type. (B) A stereoview depicts a 3D model of the S2P active site based on the placement of active site residues from a number of structurally divergent metalloenzymes (thermolysin 2tlx, mitochondrial processing peptidase 1hr8, peptide deformylase 1Bs8, and leishmanolysin 1lml). Portions of conserved TMH segments (white helixes) were modeled with an HExxH motif-containing metalloprotease helix (residues 256-270 from 1lml) and a proline-containing TMH helix (residues A18-A33 from 1xqf). Side chains of conserved S2P residues required for catalysis (black) and of other conserved S2P residues (gray) are depicted with bond representation, and the zinc ligand is illustrated with partially transparent CPK. The backbone of a peptide substrate (from 1hr8) is shown as ball-and-stick and indicates the potential relative placement of the SREBP peptide substrate.