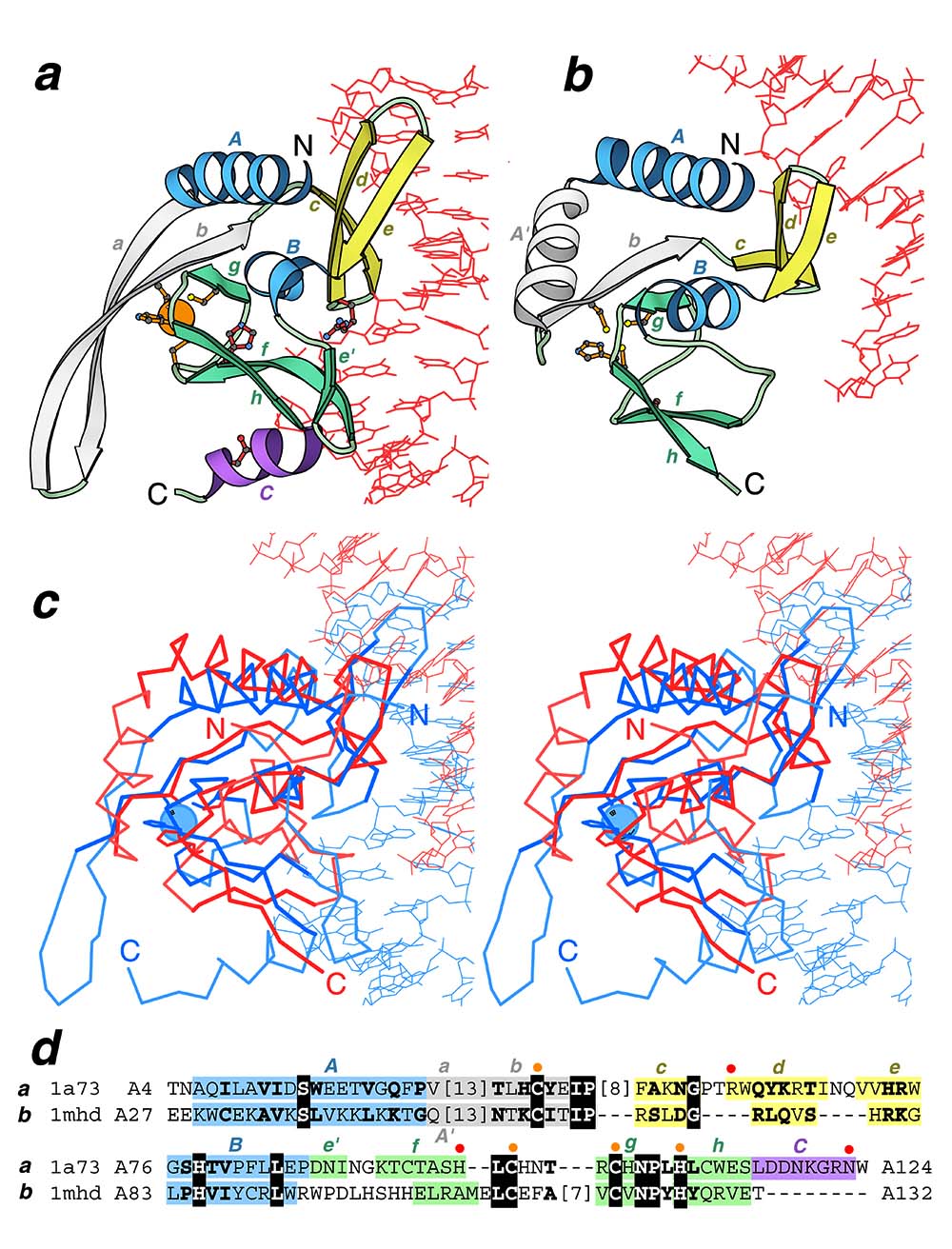
Global structural similarity between homing endonuclease and MH1 domain of Smad: Ribbon diagrams of a homing endonuclease I-PpoI from Physarum polycephalum (PDB entry 1a73, residues A7-A125) and b human Smad3 MH1 domain (PDB entry 1mhd, residues A29-A132) in complex with DNA were drawn by Bobscript, a modified version of Molscript. The structures were superimposed and then separated for clarity. N- and C-termini are labeled. The spatially equivalent structural elements are colored correspondingly in the two structures. α-Helices/β-strands in the N- and C-terminal subdomains are colored in blue/yellow, and in purple/green respectively. Insertion in the N-terminal domain is shown in gray. DNA chains are red. Side chains of active site residues (red) and zinc ligands (orange) in homing endonuclease and corresponding residues in MH1 Smad are shown in ball-and-stick presentation. Zinc ion is shown as orange ball. c Structure superposition and d structure-based sequence alignment of endonuclease I-PpoI (1a73) and Smad3 MH1 (1mhd) generated by DALI and modified manually. The panel label, PDB entry name, starting and ending residue numbers are given for each protein. Invariant residues are boxed with black and conserved substitutions are shown in bold letters. The numbers of residues omitted from the alignment are shown in brackets. Color shading and labels of secondary structure elements correspond to those in a and b. The active site residues and zinc ligands in endonuclease are marked above the alignment with red and orange dots respectively and their side chains are displayed on the panels a and b.