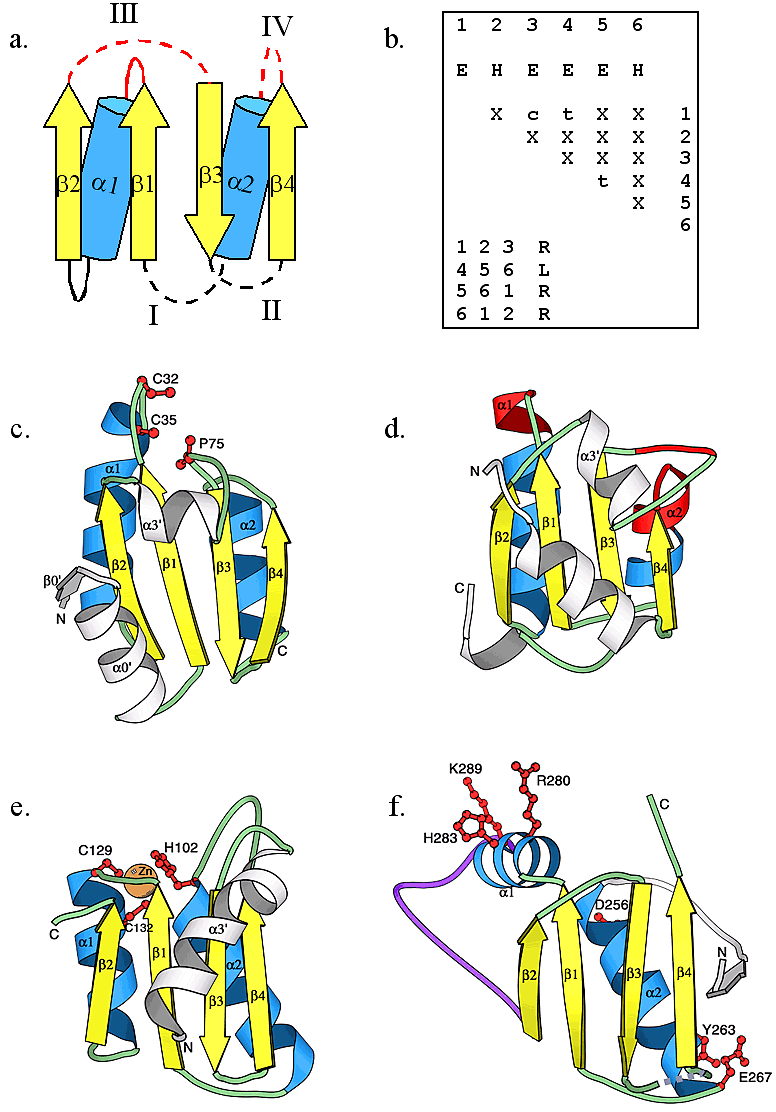
Thioredoxin fold and its observed circular permutations. (a) The topological diagram of the thioredoxin-like fold. α-helices and β-strands are shown as blue cylinders and yellow arrows, respectively, the lines connecting different secondary structures represent loop regions between them. Dotted loops indicate the termini positions of the four types of circular permutations that we observed. No termini were observed at solid loop locations. Loops shown in red indicate the common location of an active. (b) The query matrix of the thioredoxin-like fold of type I circular permutation. Secondary structures are consecutively numbered in arabic numbers. Upper case letters E (β-strand) and H (α-helix) indicate the type of secondary structure. Lower case letters c and t indicate parallel and anti-parallel hydrogen-bonding interactions between secondary structures, respectively. Upper case letter X indicates that no interactions were considered. Upper case letters R and L indicate right-handed and left-handed chirality in a triplet of secondary structures, respectively. Ribbon diagrams of (c) human thioredoxin (1ert53), a representative of type I circular permutation, (d) yeast ribosomal protein L30 (1cn8A39), a representative of type II circular permutation, (e) E. coli cytidine deaminase (1aln_148), a representative of type III circular permutation, and (f) bacteriophage HK97 capsid protein gp5 (1ohg54, previous PDB ID: 1fh6), a representative of type IV circular permutation, were produced using the program MOLSCRIPT. Corresponding secondary structure elements are colored and named as in diagram (a). Elements corresponding to inserted domains are shown in white. The long insertion in capsid protein gp5 is shown in purple in (f). In (c), (e), and (f), active site residues are depicted in red ball-and-stick representation. In (d), active site residues interacting with RNA are shown in red. The orange sphere in (e) shows a zinc ion.