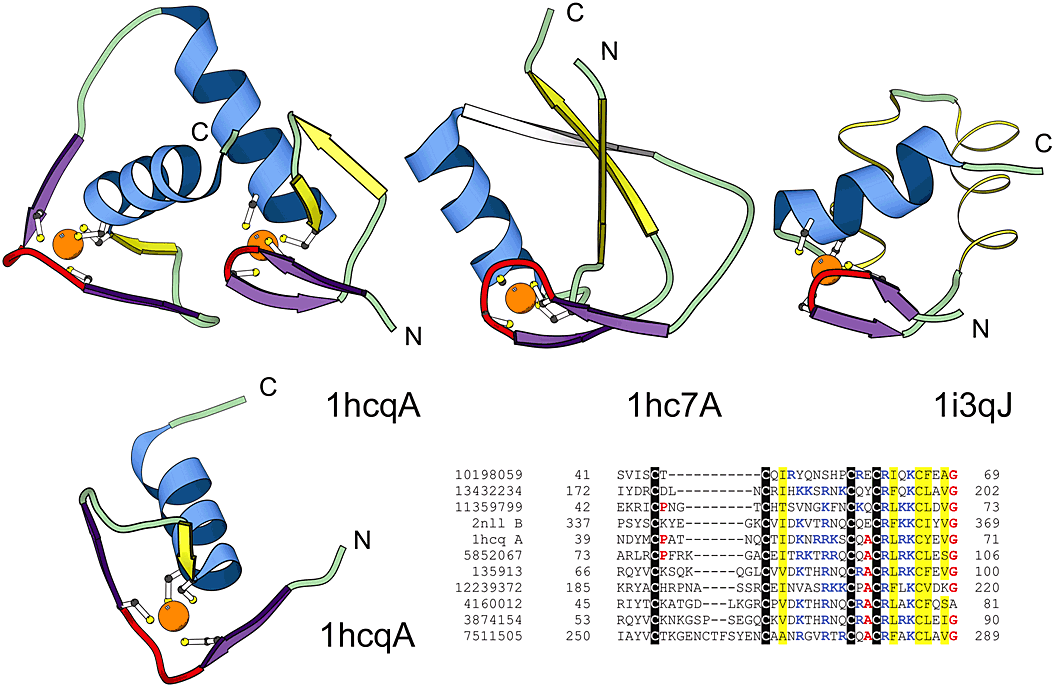
Structural comparisons of representative treble clef fingers. Structural diagrams of the treble clef of the steroid hormone "estrogen" receptor DNA-binding domain showing both zinc-binding sites (1hcq, chain A, residues 3-36 and 39-71), prolyl-tRNA synthetase (1hc7, chain A, residues 454-477; 418-440) and the RPB10 protein from RNA polymerase II (1i3q, chain J, residues 3-54) are shown to illustrate the wide range of variations in the structure of the treble clef finger. The N-terminal site of 1hcq is a typical treble clef motif. The C-terminal site (below), reoriented for clarity, lacks the β-hairpin shown in yellow for the N-terminal treble clef and has a distorted knuckle (red). This zinc-binding region shows some resemblance to other treble clef fingers in the placement of the ligands for zinc-binding. However, the knuckle, which contributes the other two ligands in treble clef fingers, is significantly different and does not align structurally with those in other fingers. PSI-BLAST alignment of representative nuclear receptor sequences is also shown. The sequences included in the alignment are: the nuclear factor NHR-63 (gi 10198059), peroxisome proliferator activated receptor (gi 13432234), NHR-18 (gi 11359799), retenoid X receptor (2nll), estrogen receptor (1hcq), tailless protein (TLL_DROME gi 135913), FTZ-F1 (gi 12239372), dissatisfaction (gi 4160012), NHR-67 (gi 3874154) and NHR-2 (gi 7511505).