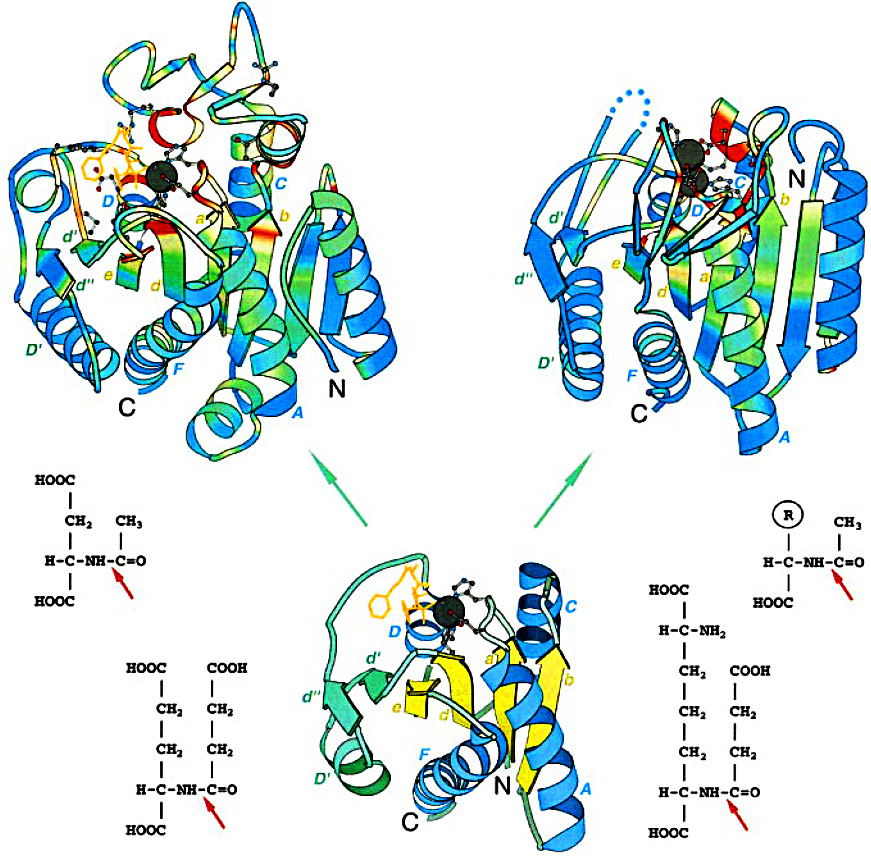
Structure-function comparison of Zn carboxypeptidase (ZnCP) and Zn aminopeptidase (ZnAP) families: Top left: carboxypeptidase A (PDB entry 8cpa); top right: an enzyme from aminopeptidase family called carboxypeptidase G2 (PDB entry 1cg2). The structures were superimposed and then separated for clarity. Ribbon diagrams are rainbow-colored by sequence conservation in a multiple sequence alignment of each family (ZnCP and ZnAP). Red corresponds to the highest conservation and blue corresponds to the lowest conservation. Dark-gray balls show zinc ions and conserved side-chains are displayed in ball-and-stick presentation. Inhibitor present in carboxypeptidase A structure is shown in orange. Dots in carboxypeptidase G2 structure stand for the not shown inserted domain of about 100 amino acid residues. N and C termini and core secondary structure elements are labeled. Bottom middle: the "minimal structure" of a possible ancestor of the superfamily. The carboxypeptidase A coordinates were used to generate the structure. Only the structural elements present in all of the sequences in the alignment of the ZnCP/AP superfamily are shown. The Rossman fold type 2 + 2 β-sheet is shown in yellow and α-helices are blue, a right-handed βαβ-unit (d'D'd") inserted between β-stand d and α-helix D is colored green. The molecules are rendered with BOBSCRIPT a modified version of MOLSCRIPT. Bottom left: succinyl-glutamate (bottom) and N-acetyl-L-aspartic acid (top), substrates for the enzymes from ZnCP family; bottom right: succinyl-diaminopimelate (bottom) and N-acetyl-L-amino acid (top), substrates for the enzymes from ZnAP family. Red arrows point at the scissile bond.