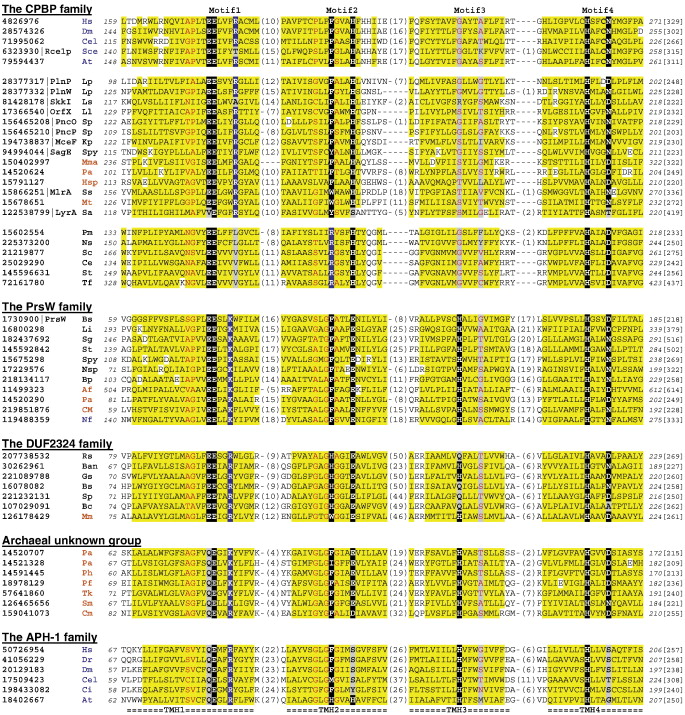
Multiple sequence alignments of representative members from protein families homologous to the type II CAAX prenyl proteases Multiple sequence alignments of representative members from protein families homologous to the type II CAAX prenyl proteases Four predicted core transmembrane helical segments (labeled TMH1-4 below the sequences) with conserved motifs (labeled Motif1-4 above the sequences) are shown for representative sequences of the CPBP family, the PrsW proteases, the DUF2324 family, a group of archaeal proteins with unknown function, and APH-1 proteins. The three separated groups in the CPBP family are eukaryotic type II CAAX proteases, prokaryotic homologs including putative bacteriocin-processing enzymes, and a group of bacterial proteins with the conserved arginine residues missing from the first motif but present in the second motif. Putative active site residues are shown on black background, except for the conserved positively charged residues, which are shown on blue background. Substitutions in these positions are on grey background. Non-charged residues in mainly hydrophobic positions are on yellow background. Small residues (residue types: G, A, S, C, V, T, N, D and P) in positions with mainly small residues are shown in red letters. NCBI gene identification numbers, along with common names for some proteins, are shown before the species name abbreviations. Numbers of residues in between the segments are shown in parentheses. Starting/ending residue numbers and sequence lengths are shown in italic font and in brackets, respectively. Species name abbreviations are: At: Arabidopsis thaliana, Af: Archaeoglobus fulgidus, Ba: Bacillus anthracis, Bs: Bacillus subtilis, Bp: Bacteroides pectinophilus, Bc: Burkholderia cenocepacia, Cel: Caenorhabditis elegans, Cm: Caldivirga maquilingensis, Ci: Ciona intestinalis, Ce: Corynebacterium efficiens, Dr: Danio rerio, Dm: Drosophila melanogaster, Gs: Geobacillus sp., Hsp: Halobacterium sp., Hs: Homo sapiens, Kp: Klebsiella pneumoniae, Lp: Lactobacillus plantarum, Ll: Lactococcus lactis, Li: Listeria innocua, Ls: Lactobacillus sakei, Mma: Methanococcus maripaludis, Mm: Methanoculleus marisnigri, Mp: Methanosphaerula palustris, Mt: Methanothermobacter thermautotrophicus, Ns: Neisseria subflava, Nf: Neosartorya fischeri, Nsp: Nostoc sp., Pm: Pasteurella multocida, Pa: Pyrococcus abyssi, Pf: Pyrococcus furiosus, Ph: Pyrococcus horikoshii, Rs: Ralstonia solanacearum, Sce: Saccharomyces cerevisiae, St: Salinispora tropica, Ss: Sphingomonas sp., Sa: Staphylococcus aureus, Sm: Staphylothermus marinus, Sp: Streptococcus pneumoniae, Spy: Streptococcus pyogenes, Sc: Streptomyces coelicolor, Sg: Streptomyces griseus, Tf: Thermobifida fusca, and Tk: Thermococcus kodakarensis. Domain color-coding: bacteria - black, archaea - orange, and eukaryotes - blue.