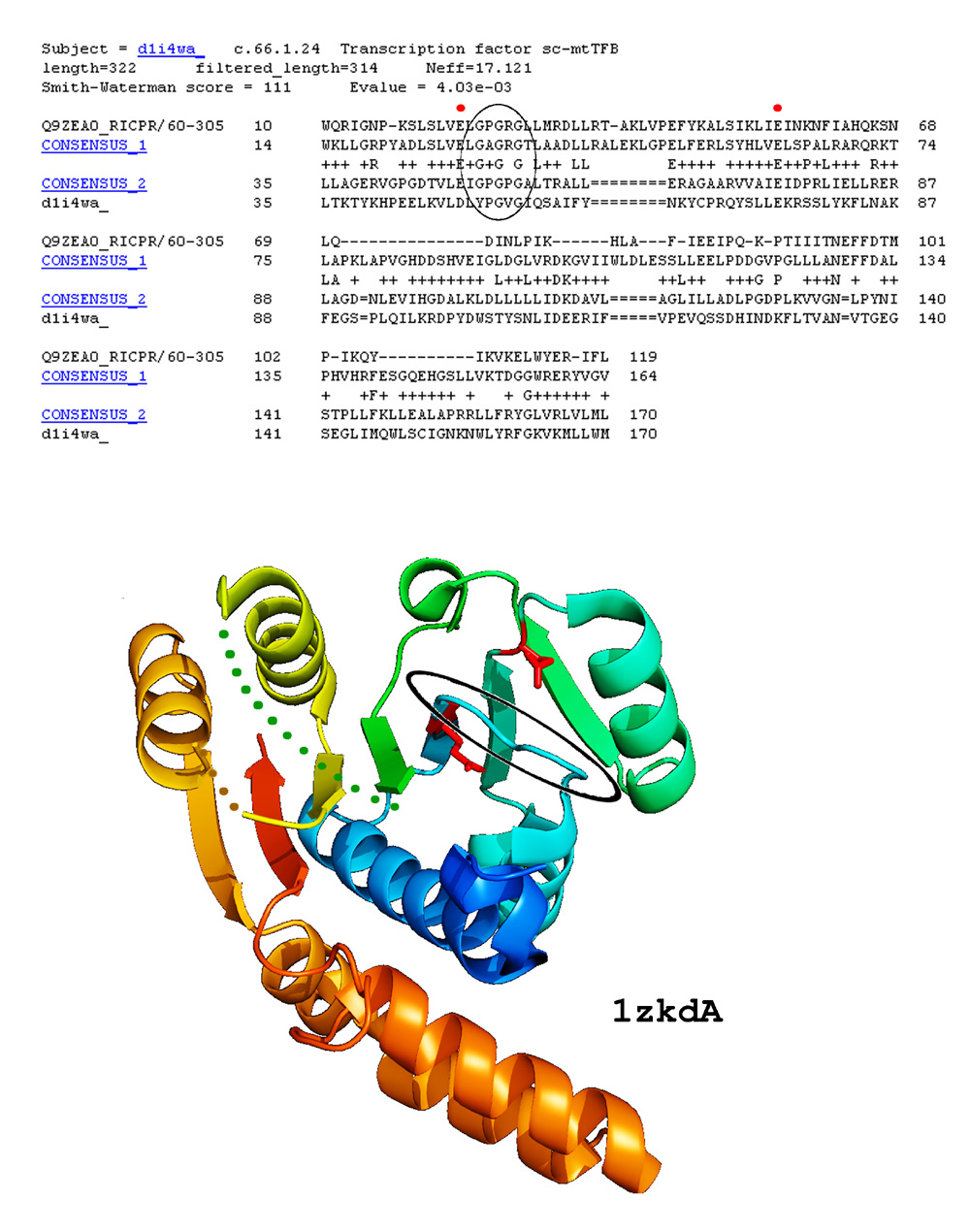
Example of COMPASS profile-profile alignment confirmed by a recently solved 3D structure: The header includes brief information about the hit, followed by COMPASS score and E-value. In this example, the top and consensus sequences for compared profiles are displayed. Position matches with positive scores are marked by '+', identical residues in the two consensus sequences are marked by the residue symbol. Invariant glutamates of Motifs I and II involved in ligand binding by S-adenosyl-l-methionine-dependent methyltransferases (SAM-Mtases) are marked with red dots, glycine-rich motif is circled. Below: A recently solved structure for a member of DUF185 family (PDB ID 1zkd) confirms our prediction. Side chains of the invariant glutamate residues are shown in red, glycine-rich loop is circled.