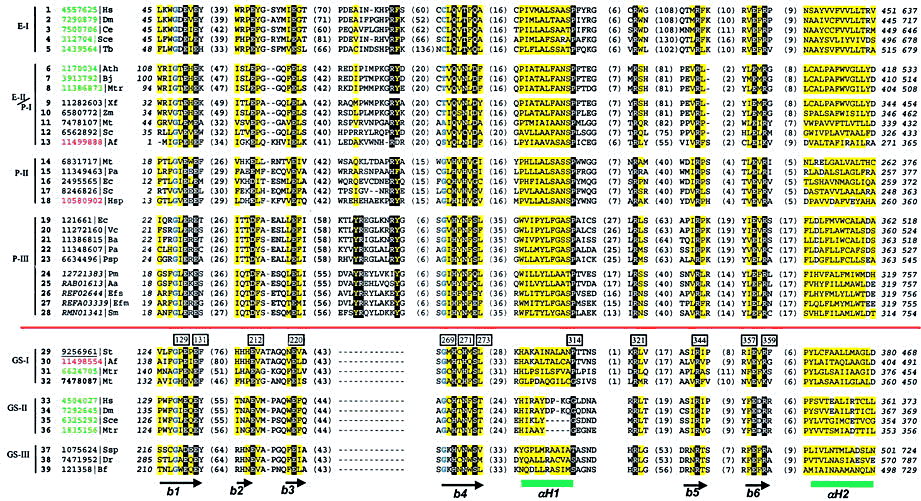
Multiple sequence alignment for representative sequences of the γ-GCS and GS superfamily: γ-GCS family sequences are above the red line. E-I represents animal/fungi sequences; E-II represents plant sequences. P-I, P-II, and P-III are prokaryotic groups. Three types of GSs (GS-I, -II, and -III) are shown below the red line. Each sequence is identified by the NCBI gene identification number (gi) except for those from the ERGO data base, which use the identifiers (italicized) in the ERGO data base. Sequence identifiers are followed by abbreviations of the species names. The archaeal and eukaryotic identifiers are marked with red and green, respectively, and the gi number of the sequence with known structure is underlined (gi|9256961, Protein Data Bank entry 1F52). The identifier of GS closest to the γ-GCS family members is shown in bold (gi|7478087). The first and the last residue numbers of the shown sequences are indicated, and the total length of the proteins is shown at the end (in italicized numbers). Only conserved motifs are shown. Residues in between the motifs are not displayed and the number of omitted residues is shown in parentheses. Uncharged residues at mainly hydrophobic positions are shaded yellow. Conserved small residues are in blue bold letters. The secondary structural elements are shown at the bottom according to the structure of S. typhimurium GS (PDB entry 1F52). β-Strands and α-helices are shown as black arrows and green rectangles, respectively. Conserved residues discussed in the text are marked by numbers (based on the S. typhimurium sequence) in the boxes above the GS sequences and those depicted on the structure diagram are shown in white bold letters on black background. The species name abbreviations are: Aa, Actinobacillus actinomycetemcomitans; Af, A. fulgidus; Ath, Arabidopsis, thiliana; Ba, Buchnera aphidicola; Bf, Bacteroides fragilis; Bj, Brassica juncea; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Deinococcus radiodurans; Ec, E. coli; Efm, Enterococcus faecium; Efs, Enterococcus faecalis; Hsp, Halobacterium sp.; Mt, Mycobacterium tuberculosis; Mtr, Medicago truncatula; Pa, Pseudomonas aeruginosa; Pm, Pasteurella multocida; Psp, Pseudomonas sp.; Sc, Streptomyces coelicolor; Sce, S. cerevisiae; Sm, Streptococcus mutans; Ssp, Synechocystis sp.; St, Salmonella typhimurium; Tb, T. brucei; Vc, Vibrio cholerae; Xf, Xylella fastidiosa; Yp, Yersinia pestis; Zm, Zymomonas mobilis.