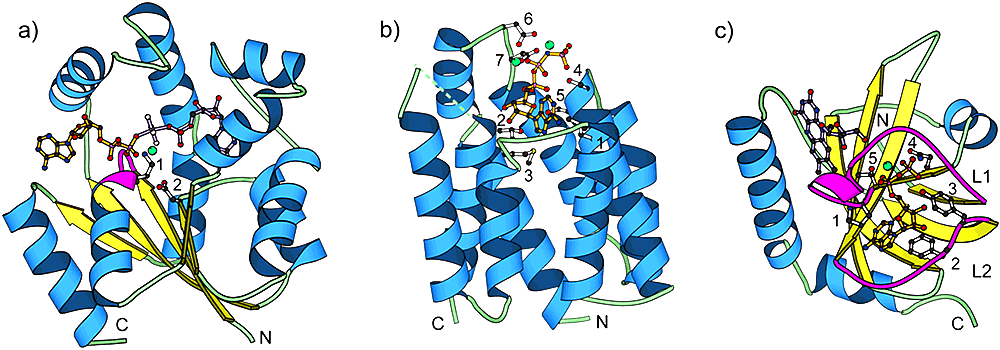
Proteins of different structure classes can be kinases. We illustrate that a given chemical reaction (e.g. transfer of a phosphate from ATP to a substrate) can be performed by completely different structure types. a) α/β – UMP/CMP kinase (PDB|1qf9), a member of the P-loop kinase family. Residue 1 and 2 are the conserved Lys19 of the Walker A motif and the conserved Asp89 of the Walker B motif, respectively. Lys19 coordinates the β- and γ-phosphates of the nucleotide, while Asp89 coordinates the magnesium cation. The P-loop is shown in magenta. b) all-α – Dihydroxyacetone kinase nucleotide-binding domain (PDB|1un9). Residues 1 (Leu435), 2 (Thr476), and 3 (Met477) pack around the adenine ring of the nucleotide. Residues 4 (Ser431) and 5 (Ser432) interact with the phosphate tail of the nucleotide. Residues 6 (Asp385) and 7 (Asp387) are involved in coordinating the two magnesium cations. c) mostly β – Riboflavin kinase (PDB|1nb9). Loops L1 and L2 are shown in magenta. Residues 1 (Pro33) and 2 (Phe97) interact with the adenine ring of the nucleotide. Residues 3 (Tyr98) and 4 (Asn36) interact with the phosphate tail of the nucleotide. Residue 5 (Thr34) coordinates the magnesium cation. In this and all other structure figures, the ATP analog is colored orange, substrate molecules are purple, and magnesium cations are green balls. Ribbon diagrams were made using the program MOLSCRIPT.