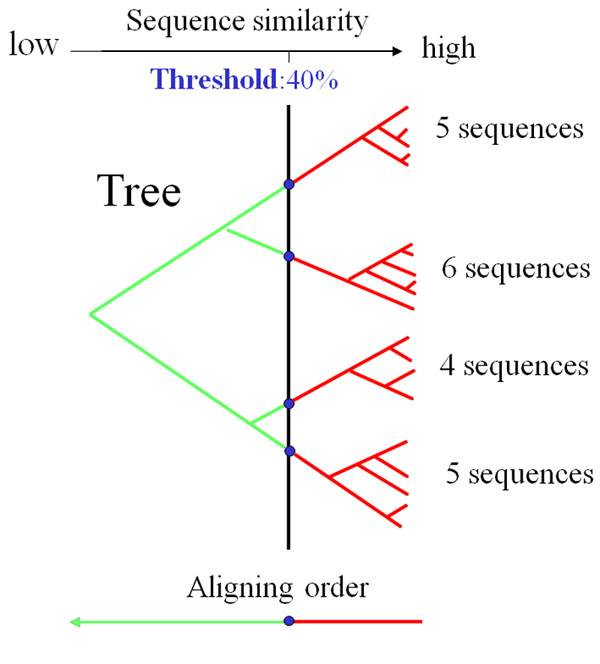
Partitioning of fast and slow alignment processes in PCMA. In this example, a tree is built for twenty input sequences. At similarity cutoff 40%, four groups of similar sequences are aligned in a fast way within each group. The alignments between the four relatively divergent groups are constructed by a profile consistency-based method that is slower but more accurate. Using a fast alignment strategy on easy cases (highly similar sequences) an a more elaborate strategy on difficult cases (divergent sequences) balances alignment speed and accuracy. This idea is also used in our other multiple alignment programs (MUMMALS, PROMALS and PROMALS3D).