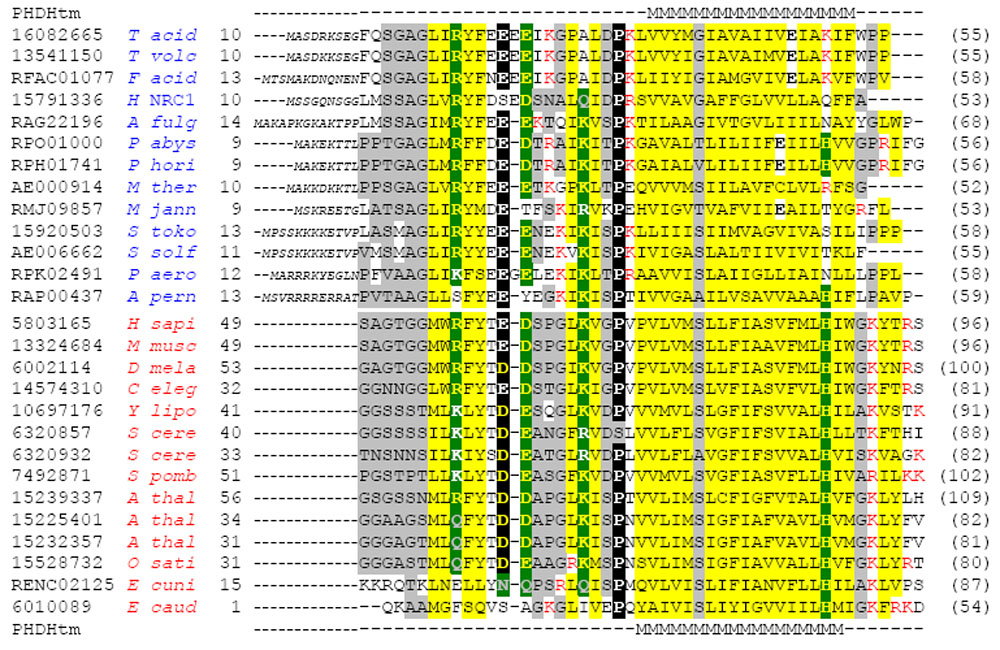
Sec61beta Multiple Sequence Alignment. Sequences are labelled according to NCBI gene identification number (gi), ERGO database identifier or GenBank identifiers (AE000914 and AE006662). Names of archaeal and eukaryotic sequences are coloured blue and red, respectively. The sequential number of the first amino acid in the motif is indicated preceding the sequence, and the sequence length is indicated in parentheses following the sequence. Complete archaeal sequences are shown. Amino acids that precede the motif and do not align with residues from the longer eukaryotic sequences (not shown) are in small italicized letters. Residues conserved across eukaryotic sequences and archaeal sequences are highlighted using different colour backgrounds: small (grey), hydrophobic (yellow), highly conserved (black), relatively conserved polar residues (green). Red letters indicate positively charged residues N- and C-terminal to the predicted transmembrane a helix; black letters indicate negatively charged residues in the predicted transmembrane a helix. The transmembrane predictions (PHDHtm) produced using PHD with archaeal (RPO01000) and eukaryotic (gi5803165) representative sequences as input are indicated above and below the respective alignments, with predicted membrane-spanning residues specified (M). Eukaryotic sequences were detected using PSI-BLAST (E-value cutoff 0.01, default parameters) against the non-redundant (nr) database (Nov 21, 2001; 799 241 sequences). Archaeal sequences were detected using PSI-BLAST against the nr database (E-value cutoff 0.05, BLOSUM80 matrix, default parameters), BLAST against the ERGO database (E-value cutoff 0.05), and tBLASTn against the remaining completely sequenced archaeal genome nucleotide databases. A position-specific scoring matrix (-B option in blastpgp) was generated with an alignment of all archaeal sequences and used in PSI-BLAST searches (BLOSUM80 matrix, default parameters) against the nr database to establish a link to the eukaryotic sequences. Several duplicate sequences are present in the ERGO and NCBI databases, with slight differences in the predicted start and termination sites: A. thal gi15225401 and gi13878103, A. pern RAP00437 and gi14600867, A. fulg RAG22196 and gi11499365, and T. volv gi13541150 and gi14324537. Of these sequence pairs, we include the sequence most closely resembling the remaining sequences. Species abbreviations: A. fulg, Archaeoglobus fulgidus; A. pern, Aeropyrum pernix; A. thal, Arabidopsis thaliana; C. eleg, Caenorhabditis elegans; D. mela, Drosophila melanogaster; E. caud, Entodinium caudatum; E. cuni, Encephalitozoon cuniculi; F. acid, Ferroplasma acidarmanus; H. NRC1, Halobacterium sp. NRC-1; H. sapi, Homo sapiens; M. jann, Methanococcus jannaschii; M. musc, Mus musculus; M. ther, Methanobacterium thermoautotrophicum; O. sati, Oryza sativa; P. abys, Pyrococcus abyssii; P. aero, Pyrobaculum aerophilum; P. hori, Pyrococcus horikoshii; S. cere, Saccharomyces cerevisiae; S. pomb, Schizosaccharomyces pombe; S. solf, Sulfolobus solfataricus; S. toko, Sulfolobus tokodaii; T. acid, Thermoplasma acidophilum; T. volc, Thermoplasma volcanium; Y. lipo, Yarrowia lipolytica.