| UID: | 000356629 |
|---|---|
| ID: | e3etrA2 |
| Source structure type: | experimental structure |
| ECOD representative type: | Automatic domain |
| Parent: | e1wriA1 |
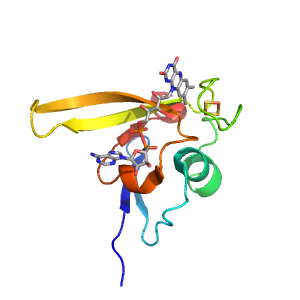
| Range: | A:2-92 |
| DrugDomain: | P80457_FAD P80457_FES | PDB: | 3etr |
| PDB Description: | Xanthine dehydrogenase/oxidase |
Structure of domain e3etrA2
Domains in the same protein:
| e3etrA1 | A:93-165 | CO dehydrogenase ISP C-domain like | CO dehydrogenase ISP C-domain like | CO dehydrogenase ISP C-domain like |
| e3etrA2 | A:2-92 | beta-Grasp | Ubiquitin-related | Ubiquitin-like |
Domains in the same PDB: