| UID: | 000424379 |
|---|---|
| ID: | e3nvkE1 |
| Source structure type: | experimental structure |
| ECOD representative type: | Automatic domain |
| Parent: | e1xbiA1 |
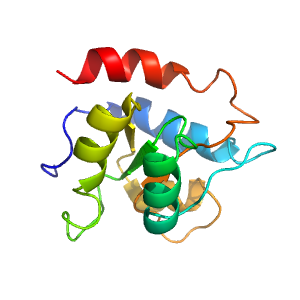
| Range: | E:4-124 | PDB: | 3nvk |
| PDB Description: | 50S ribosomal protein L7Ae |
Structure of domain e3nvkE1
Domains in the same protein:
| e3nvkE1 | E:4-124 | Bacillus chorismate mutase-like | L30e-like | L30e-like |
Domains in the same PDB: