| UID: | 002391101 |
|---|---|
| ID: | e6chgC1 |
| Source structure type: | experimental structure |
| ECOD representative type: | Automatic domain |
| Parent: | e1ml9A2 |
| Range: | C:848-1000 |
| DrugDomain: | Q6CIT4_SAM Q6CIT4_ZN Q6CLY5_SAM | PDB: | 6chg |
| PDB Description: | Histone-lysine N-methyltransferase, H3 lysine-4 specific |
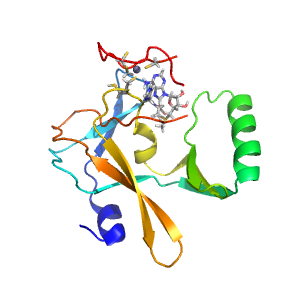
Structure of domain e6chgC1
Domains in the same protein:
| e6chgC1 | C:848-1000 | beta-clip | SET domain-like | SET domain-like |
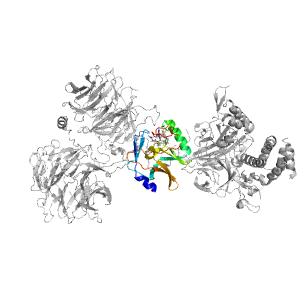
Domains in the same PDB: