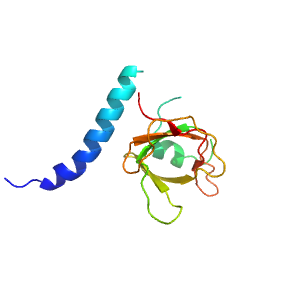
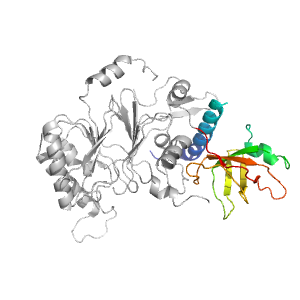
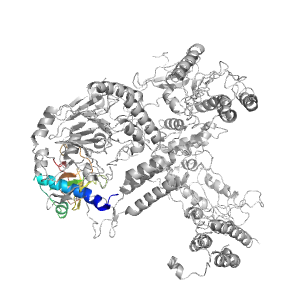
Structure of domain e6hv8B1
Domains in the same protein:
| e6hv8B2 | B:170-203,B:349-556,B:598-688 | Carbon-nitrogen hydrolase-like | Metallo-dependent phosphatases | Metallo-dependent phosphatases |
| e6hv8B1 | B:204-233,B:266-348 | OB-fold | Nucleic acid-binding protein | Nucleic acid-binding protein |
Domains in the same PDB: