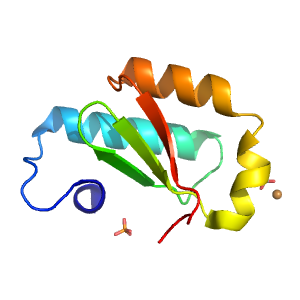
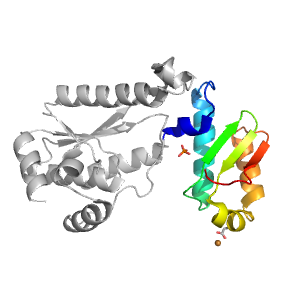
- A: a+b two layers
- X: Alpha-lytic protease prodomain-like
- H: GMP synthetase C-terminal dimerisation domain
- T: GMP synthetase C-terminal dimerisation domain
- F:
| UID: | 002549682 |
|---|---|
| ID: | e6utrD1 |
| Source structure type: | experimental structure |
| ECOD representative type: | Automatic domain |
| Parent: | e5udrF2 |
| Range: | D:177-259 |
| DrugDomain: | A0A0G9FES3_CU A0A0G9FES3_PO4 | PDB: | 6utr |
| PDB Description: | ATP-dependent sacrificial sulfur transferase LarE |