- A: a+b complex topology
- X: Protein kinase/SAICAR synthase/ATP-grasp
- H: Protein kinase/SAICAR synthase/ATP-grasp
- T: ATP-grasp
- F: CPSase_L_D2
| UID: | 000080665 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2j9gA1 |
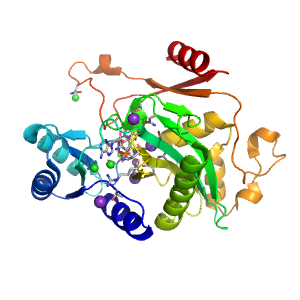
| Range: | A:128-402 |
| Ligands: | ADP CL K MN PO4 |
| PDB: | 1cs0 |
| PDB Description: | CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT |
| UniProt: | P00968 |
| Hsap BLAST neighbor: | P27708 |
| Species: | Escherichia coli |