| UID: | 000103023 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1dnpA2 |
| Range: | B:1-200 |
| Ligands: | MHF |
| PDB: | 1dnp |
| PDB Description: | DNA PHOTOLYASE |
| UniProt: | P00914 |
| Species: | Escherichia coli |
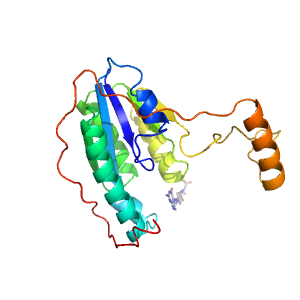
Structure of domain e1dnpB2
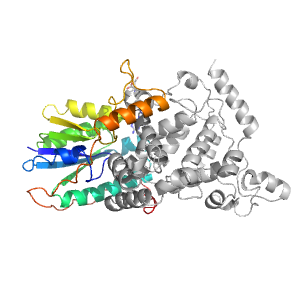
Domains in the same chain:
Domains in the same PDB:
Structural contacts of domain e1dnpB2 of H-group "HUP domains":
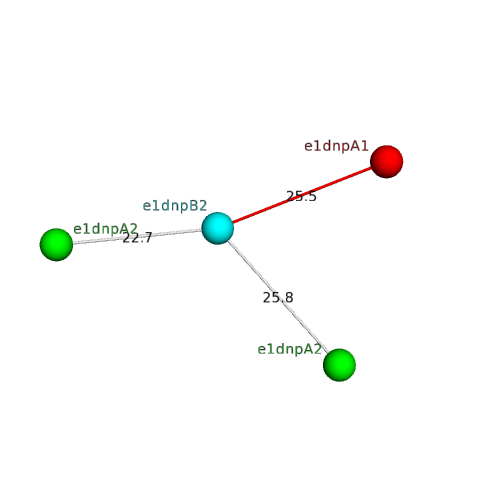
The domain and its neighboring domains (both within the asymmetric unit and to crystallographic symmetry mates) are represented by spheres and linked by lines. Distances between the center of the domain and interfaces are shown for each contact.
| Domain ID | Symmetry operator | H-group | Visualization |
|---|---|---|---|
| e1dnpA1 | B:x,y,z->A:X,Y,Z | Cryptochrome/photolyase FAD-binding domain-related | Interaction Interface Pymol |
| e1dnpA2 | B:x,y,z->A:X-1,Y+1,Z-1 | HUP domains | Interaction Interface Pymol |
| e1dnpA1 | B:X,Y-1,Z->A:x,y,z | Cryptochrome/photolyase FAD-binding domain-related | Interaction Interface Pymol |
| e1dnpA2 | B:x,y,z->A:X-1,Y,Z-1 | HUP domains | Interaction Interface Pymol |
| e1dnpA1 | B:x,y,z->A:X,Y,Z-1 | Cryptochrome/photolyase FAD-binding domain-related | Interaction Interface Pymol |