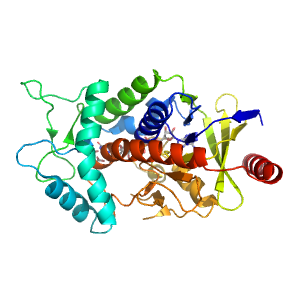
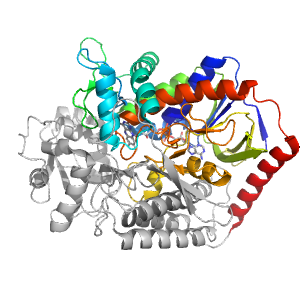
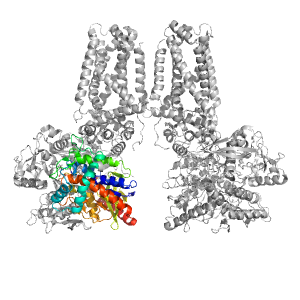
- A: a/b three-layered sandwiches
- X: Rossmann-like
- H: Rossmann-related
- T: FAD/NAD(P)-binding domain
- F: FAD_binding_2
| UID: | 000098858 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1kf6A3 |
| Range: | A:0-225,A:358-442 |
| Ligands: | FAD OAA |
| PDB: | 1l0v |
| PDB Description: | FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT |
| UniProt: | P00363 |
| Hsap BLAST neighbor: | P31040 |
| Species: | Escherichia coli |