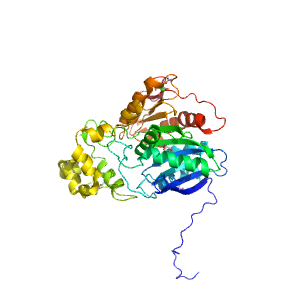
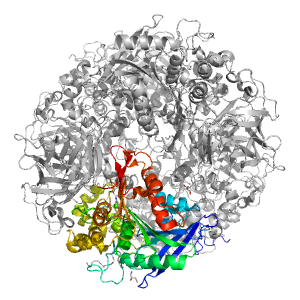
- A: a/b three-layered sandwiches
- X: alpha/beta-Hydrolases
- H: alpha/beta-Hydrolases
- T: alpha/beta-Hydrolases
- F: Peptidase_S15
| UID: | 000112160 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1ju3A2 |
| Range: | C:24-404 |
| Ligands: | CA GOL |
| PDB: | 1mpx |
| PDB Description: | alpha-amino acid ester hydrolase |
| UniProt: | Q6YBS3 |
| Species: | Xanthomonas citri |