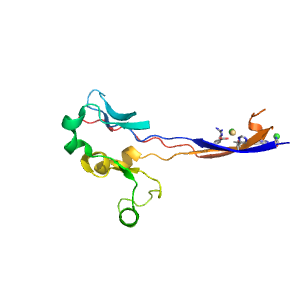
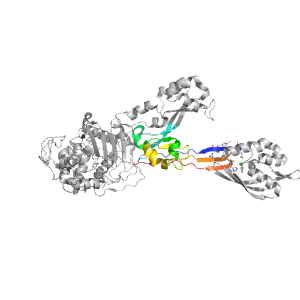
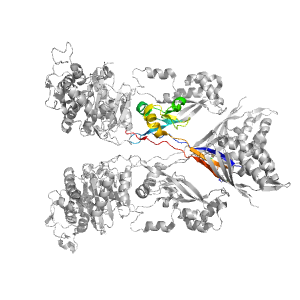
- A: beta complex topology
- X: Penicillin binding protein dimerisation domain
- H: Penicillin binding protein dimerisation domain
- T: Penicillin binding protein dimerisation domain
- F: PBP_dimer
| UID: | 001394012 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1rp5A6 |
| Range: | B:139-167,B:239-327 |
| Ligands: | CD CL |
| PDB: | 1mwr |
| PDB Description: | PENICILLIN-BINDING PROTEIN 2A |
| UniProt: | Q93IC2 |
| Species: | Staphylococcus aureus |