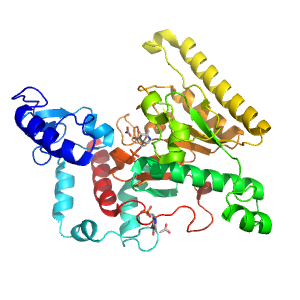
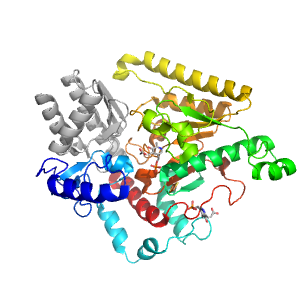
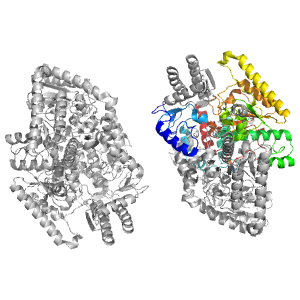
- A: a/b three-layered sandwiches
- X: PLP-dependent transferases
- H: PLP-dependent transferases
- T: PLP-dependent transferases
- F: Aminotran_3
| UID: | 001151794 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1zodA3 |
| Range: | C:11-375 |
| Ligands: | ACT FES PLP |
| PDB: | 1ohv |
| PDB Description: | 4-AMINOBUTYRATE AMINOTRANSFERASE |
| UniProt: | P80147 |
| Hsap BLAST neighbor: | P80404 |
| Species: | Sus scrofa |