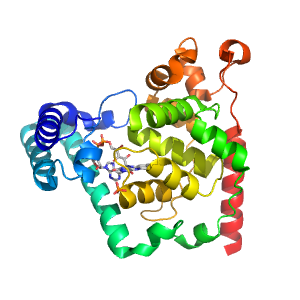
- A: alpha complex topology
- X: Cryptochrome/photolyase FAD-binding domain-related
- H: Cryptochrome/photolyase FAD-binding domain-related
- T: Cryptochrome/photolyase FAD-binding domain
- F: FAD_binding_7
| UID: | 000061968 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2j07A1 |
| Range: | A:205-475 |
| Ligands: | FAD PO4 |
| PDB: | 1owm |
| PDB Description: | Deoxyribodipyrimidine photolyase |
| UniProt: | P05327 |
| Hsap BLAST neighbor: | Q49AN0 |
| Species: |