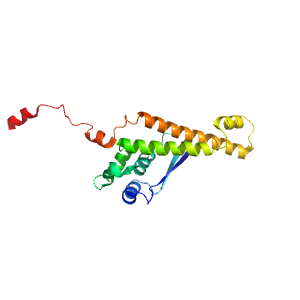
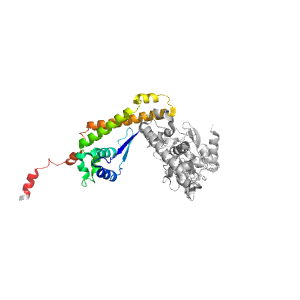
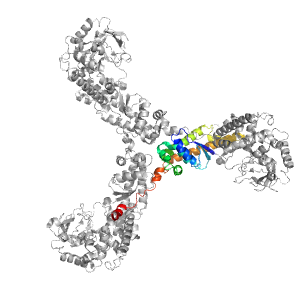
- A: a+b complex topology
- X: RuBisCo LSMT C-terminal, substrate-binding domain
- H: RuBisCo LSMT C-terminal, substrate-binding domain
- T: RuBisCo LSMT C-terminal, substrate-binding domain
- F: Rubis-subs-bind
| UID: | 000085502 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2h2jA2 |
| Range: | C:311-486 |
| PDB: | 1ozv |
| PDB Description: | Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast |
| UniProt: | Q43088 |
| Hsap BLAST neighbor: | Q86TU7 |
| Species: | Pisum sativum |