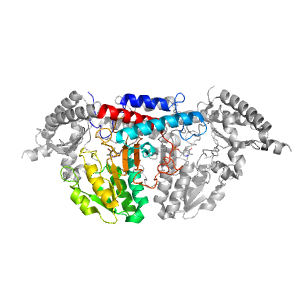
- A: a/b three-layered sandwiches
- X: PLP-dependent transferases
- H: PLP-dependent transferases
- T: PLP-dependent transferases
- F: Aminotran_5
| UID: | 001152488 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1eg5A2 |
| Range: | B:2-263 |
| Ligands: | PLP |
| PDB: | 1p3w |
| PDB Description: | Cysteine desulfurase |
| UniProt: | P0A6B7 |
| Hsap BLAST neighbor: | Q9Y697 |
| Species: | Escherichia coli |