- A: a/b three-layered sandwiches
- X: Rossmann-like
- H: Rossmann-related
- T: DHS-like NAD/FAD-binding domain
- F: TPP_enzyme_M
| UID: | 000099230 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2ez9A1 |
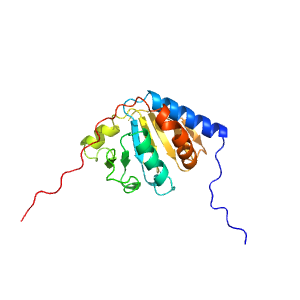
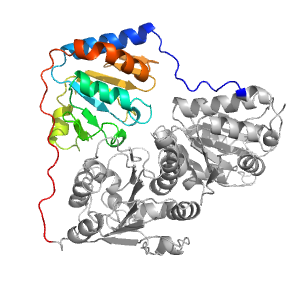
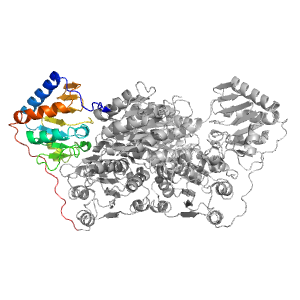
| Range: | B:182-360 |
| PDB: | 1pyd |
| PDB Description: | PYRUVATE DECARBOXYLASE |
| UniProt: | P06169 |
| Species: | Saccharomyces cerevisiae |