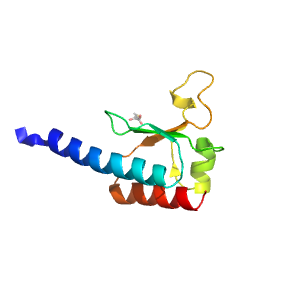
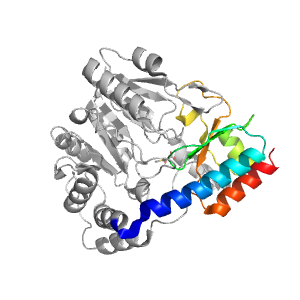
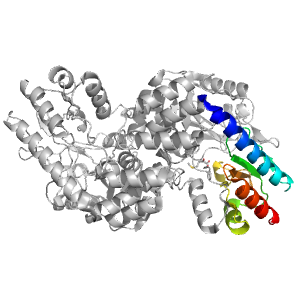
- A: a+b two layers
- X: C-terminal domain in some PLP-dependent transferases
- H: C-terminal domain in some PLP-dependent transferases
- T: C-terminal domain in some PLP-dependent transferases
- F: Aminotran_1_2
| UID: | 001151834 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1m7yA1 |
| Range: | A:272-368 |
| Ligands: | KMT |
| PDB: | 1v2e |
| PDB Description: | GLUTAMINE AMINOTRANSFERASE |
| UniProt: | Q75WK2 |
| Species: | Thermus thermophilus |