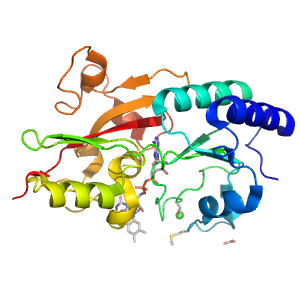
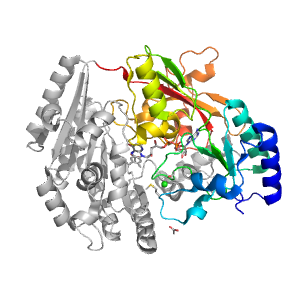
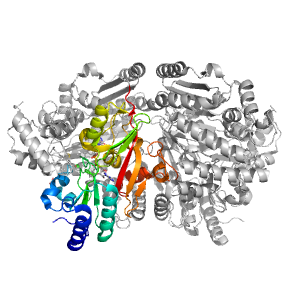
- A: a+b complex topology
- X: FAD-binding domain-like
- H: FAD-binding domain
- T: FAD-binding domain
- F: FAD_binding_4
| UID: | 000080832 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1wvfA2 |
| Range: | A:7-242 |
| Ligands: | ACY CL FAD |
| PDB: | 1wve |
| PDB Description: | 4-CRESOL DEHYDROGENASE [HYDROXYLATING] FLAVOPROTEIN SUBUNIT |
| UniProt: | P09788 |
| Hsap BLAST neighbor: | Q86WU2 |
| Species: | Pseudomonas putida |