- A: a/b three-layered sandwiches
- X: The "swivelling" beta/beta/alpha domains
- H: The "swivelling" beta/beta/alpha domain
- T: The "swivelling" beta/beta/alpha domain
- F: PEP-utilizers
| UID: | 000011426 |
|---|---|
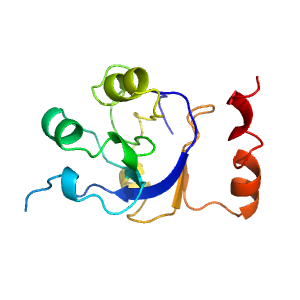
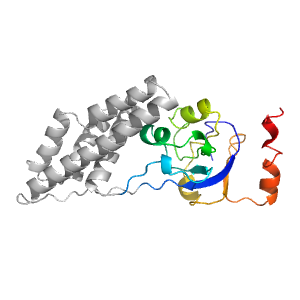
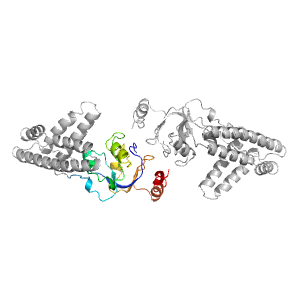
| Type: | Manual and representative domain |
| Range: | A:3-21,A:145-249 |
| PDB: | 1zym |
| PDB Description: | ENZYME I |
| UniProt: | P08839 |
| Species: | Escherichia coli |