- A: a/b three-layered sandwiches
- X: Rossmann-like
- H: Rossmann-related
- T: FAD/NAD(P)-binding domain
- F: Pyr_redox_2
| UID: | 001270538 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1gesA3 |
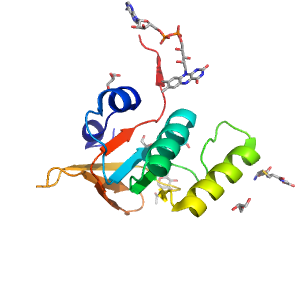
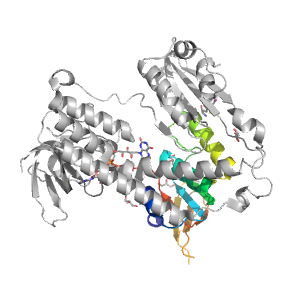
| Range: | A:177-299 |
| Ligands: | FAD GOL GSH NAG |
| PDB: | 2hqm |
| PDB Description: | GLUTATHIONE REDUCTASE |
| UniProt: | P41921 |
| Hsap BLAST neighbor: | P00390 |
| Species: | Saccharomyces cerevisiae |