- A: a+b two layers
- X: FAD-linked reductases, C-terminal domain-like
- H: FAD/NAD-linked reduatases, dimerisation (C-terminal) domain
- T: FAD/NAD-linked reduatases, dimerisation (C-terminal) domain
- F: Pyr_redox_dim
| UID: | 001197297 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1gesA1 |
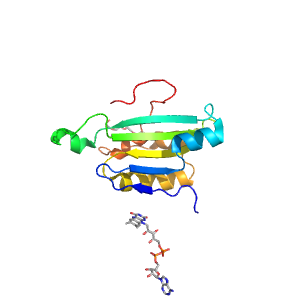
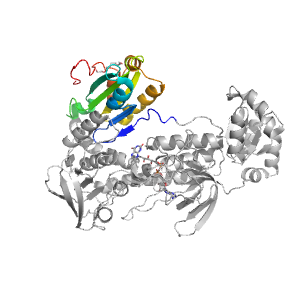
| Range: | A:465-593 |
| Ligands: | FAD PG4 |
| PDB: | 2v6o |
| PDB Description: | THIOREDOXIN GLUTATHIONE REDUCTASE |
| UniProt: | Q962Y6 |
| Hsap BLAST neighbor: | Q16881 |
| Species: | Schistosoma mansoni |