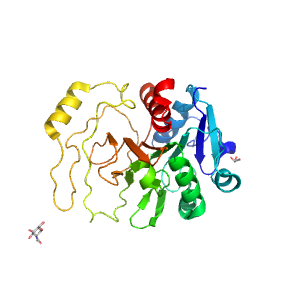
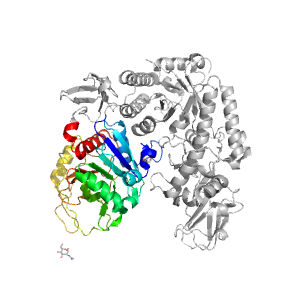
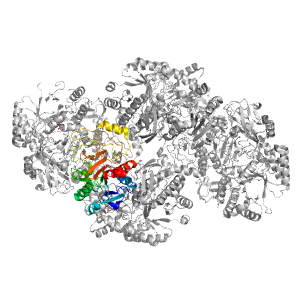
- A: a/b three-layered sandwiches
- X: Flavodoxin-like
- H: Class I glutamine amidotransferase-like
- T: Class I glutamine amidotransferase-like
- F: LBP_M
| UID: | 001837896 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2zuuB1 |
| Range: | C:437-694 |
| Ligands: | GOL NDG |
| PDB: | 2zuw |
| PDB Description: | Lacto-N-biose phosphorylase |
| UniProt: | E8MF13 |
| Species: | Bifidobacterium longum |